The landscape of viral expression and host gene fusion and adaptation in human cancer
- PMID: 24085110
- PMCID: PMC3806554
- DOI: 10.1038/ncomms3513
The landscape of viral expression and host gene fusion and adaptation in human cancer
Abstract
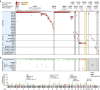
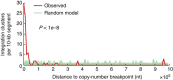
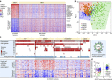
Viruses cause 10-15% of all human cancers. Massively parallel sequencing has recently proved effective for uncovering novel viruses and virus-tumour associations, but this approach has not yet been applied to comprehensive patient cohorts. Here we screen a diverse landscape of human cancer, encompassing 4,433 tumours and 19 cancer types, for known and novel expressed viruses based on >700 billion transcriptome sequencing reads from The Cancer Genome Atlas Research Network. The resulting map confirms and extends current knowledge. We observe recurrent fusion events, including human papillomavirus insertions in RAD51B and ERBB2. Patterns of coadaptation between host and viral gene expression give clues to papillomavirus oncogene function. Importantly, our analysis argues strongly against viral aetiology in several cancers where this has frequently been proposed. We provide a virus-tumour map of unprecedented scale that constitutes a reference for future studies of tumour-associated viruses using transcriptome sequencing data.
Figures



References
-
- Williams R. Global challenges in liver disease. Hepatology 44, 521–526 (2006). - PubMed
-
- Strong K., Mathers C., Epping-Jordan J., Resnikoff S. & Ullrich A. Preventing cancer through tobacco and infection control: how many lives can we save in the next 10 years? Eur. J. Cancer Prev. 17, 153–161 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

