Integrated glycoproteomics demonstrates fucosylated serum paraoxonase 1 alterations in small cell lung cancer
- PMID: 24085812
- PMCID: PMC3879622
- DOI: 10.1074/mcp.M113.028621
Integrated glycoproteomics demonstrates fucosylated serum paraoxonase 1 alterations in small cell lung cancer
Abstract
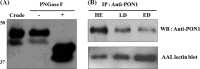
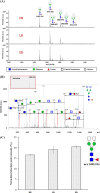
Small cell lung cancer (SCLC) is an aggressive type of lung cancer, and the detection of SCLCs at an early stage is necessary for successful therapy and for improving cancer survival rates. Fucosylation is one of the most common glycosylation-based modifications. Increased levels of fucosylation have been reported in a number of pathological conditions, including cancers. In this study, we aimed to identify and validate the aberrant and selective fucosylated glycoproteins in the sera of patients with SCLC. Fucosylated glycoproteins were enriched by the Aleuria aurantia lectin column after serum albumin and IgG depletion. In a narrowed down and comparative data analysis of both label-free proteomics and isobaric peptide-tagging chemistry iTRAQ approaches, the fucosylated glycoproteins were identified as up- or down-regulated in the sera of limited disease and extensive disease stage patients with SCLC. Verification was performed by multiple reaction monitoring-mass spectrometry to select reliable markers. Four fucosylated proteins, APCS, C9, SERPINA4, and PON1, were selected and subsequently validated by hybrid A. aurantia lectin ELISA (HLE) and Western blotting. Compared with Western blotting, the HLE analysis of these four proteins produced more optimal diagnostic values for SCLC. The PON1 protein levels were significantly reduced in the sera of patients with SCLC, whereas the fucosylation levels of PON1 were significantly increased. Fucosylated PON1 exhibited an area under curve of 0.91 for the extensive disease stage by HLE, whereas the PON1 protein levels produced an area under curve of 0.82 by Western blot. The glycan structural analysis of PON1 by MS/MS identified a biantennary fucosylated glycan modification consisting of a core + 2HexNAc + 1Fuc at increased levels in the sera of patients with SCLC. In addition, the PON1 levels were decreased in the sera of the Lewis lung carcinoma lung cancer mouse model that we examined. Our data suggest that fucosylated protein biomarkers, such as PON1, and their fucosylation levels and patterns can serve as diagnostic and prognostic serological markers for SCLC.
Figures





References
-
- Carney D. N., Gazdar A. F., Nau M., Minna J. D. (1985) Biological heterogeneity of small cell lung cancer. Semin. Oncol. 12, 289–303 - PubMed
-
- Shimosato Y., Nakajima T., Hirohashi S., Morinaga S., Terasaki T., Yamaguchi K., Saijo N., Suemasu K. (1986) Biological, pathological and clinical features of small cell lung cancer. Cancer Lett. 33, 241–258 - PubMed
-
- Parkin D. M., Bray F., Ferlay J., Pisani P. (2005) Global cancer statistics, 2002. CA Cancer J. Clin. 55, 74–108 - PubMed
-
- Chen L. C., Travis W. D., Krug L. M. (2006) Pulmonary neuroendocrine tumors: What (little) do we know? J. Natl. Compr. Canc. Netw. 4, 623–630 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

