Neurospora crassa, a model system for epigenetics research
- PMID: 24086046
- PMCID: PMC3783048
- DOI: 10.1101/cshperspect.a017921
Neurospora crassa, a model system for epigenetics research
Abstract
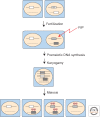
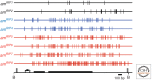
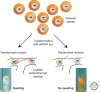
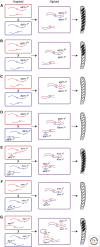
The filamentous fungus Neurospora crassa has provided a rich source of knowledge on epigenetic phenomena that would have been difficult or impossible to gain from other systems. Neurospora sports features found in higher eukaryotes but absent in both budding and fission yeast, including DNA methylation and H3K27 methylation, and also has distinct RNA interference (RNAi)-based silencing mechanisms operating in mitotic and meiotic cells. This has provided an unexpected wealth of information on gene silencing systems. One silencing mechanism, named repeat-induced point mutation (RIP), has both epigenetic and genetic aspects and provided the first example of a homology-based genome defense system. A second silencing mechanism, named quelling, is an RNAi-based mechanism that results in silencing of transgenes and their native homologs. A third, named meiotic silencing, is also RNAi-based but is distinct from quelling in its time of action, targets, and apparent purpose.
Figures




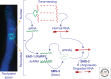
References
-
- Alexander WG, Raju NB, Xiao H, Hammond TM, Perdue TD, Metzenberg RL, Pukkila PJ, Shiu PK 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet Biol 45: 719–727 - PubMed
-
- Aramayo R, Metzenberg RL 1996. Meiotic transvection in fungi. Cell 86: 103–113 - PubMed
WWW RESOURCES
-
- http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html A website housed by the Broad Institute that serves as the official repository of the information generated by, and associated with, the Neurospora crassa genome
-
- http://www.broadinstitute.org/igv A website housed by the Broad Institute that serves as the official repository of the Integrative Genomics Viewer, a powerful Java-based high-performance visualization tool for interactive exploration of large, integrated genomic data sets
-
- http://www.repeatmasker.org A website housed by the Institute for Systems Biology that serves as the official repository for RepeatMasker, an industry-standard program used for the screening of DNA sequences for interspersed repeats and low-complexity DNA sequences
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
