Tunable stochastic pulsing in the Escherichia coli multiple antibiotic resistance network from interlinked positive and negative feedback loops
- PMID: 24086119
- PMCID: PMC3784492
- DOI: 10.1371/journal.pcbi.1003229
Tunable stochastic pulsing in the Escherichia coli multiple antibiotic resistance network from interlinked positive and negative feedback loops
Abstract
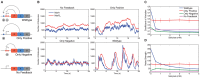
Cells live in uncertain, dynamic environments and have many mechanisms for sensing and responding to changes in their surroundings. However, sudden fluctuations in the environment can be catastrophic to a population if it relies solely on sensory responses, which have a delay associated with them. Cells can reconcile these effects by using a tunable stochastic response, where in the absence of a stressor they create phenotypic diversity within an isogenic population, but use a deterministic response when stressors are sensed. Here, we develop a stochastic model of the multiple antibiotic resistance network of Escherichia coli and show that it can produce tunable stochastic pulses in the activator MarA. In particular, we show that a combination of interlinked positive and negative feedback loops plays an important role in setting the dynamics of the stochastic pulses. Negative feedback produces a pulsatile response that is tunable, while positive feedback serves to amplify the effect. Our simulations show that the uninduced native network is in a parameter regime that is of low cost to the cell (taxing resistance mechanisms are expressed infrequently) and also elevated noise strength (phenotypic variability is high). The stochastic pulsing can be tuned by MarA induction such that variability is decreased once stresses are sensed, avoiding the detrimental effects of noise when an optimal MarA concentration is needed. We further show that variability in the expression of MarA can act as a bet hedging mechanism, allowing for survival in time-varying stress environments, however this effect is tunable to allow for a fully induced, deterministic response in the presence of a stressor.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Alekshun MN, Levy SB (2007) Molecular mechanisms of antibacterial multidrug resistance. Cell 128: 1037–1050. - PubMed
-
- Levin BR, Rozen DE (2006) Opinion - Non-inherited antibiotic resistance. Nature Reviews Microbiology 4: 556–562. - PubMed
-
- Lewis K (2010) Persister cells. Annual Review Of Microbiology 64: 357–372. - PubMed
-
- Rosner JL, Dangi B, Gronenborn AM, Martin RG (2002) Posttranscriptional Activation of the Transcriptional Activator Rob by Dipyridyl in Escherichia coli Posttranscriptional Activation of the Transcriptional Activator Rob by Dipyridyl in Escherichia coli. Journal of bacteriology 184: 1407–1416. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

