Rapid intrahost evolution of human cytomegalovirus is shaped by demography and positive selection
- PMID: 24086142
- PMCID: PMC3784496
- DOI: 10.1371/journal.pgen.1003735
Rapid intrahost evolution of human cytomegalovirus is shaped by demography and positive selection
Abstract
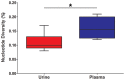
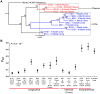
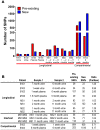
Populations of human cytomegalovirus (HCMV), a large DNA virus, are highly polymorphic in patient samples, which may allow for rapid evolution within human hosts. To understand HCMV evolution, longitudinally sampled genomic populations from the urine and plasma of 5 infants with symptomatic congenital HCMV infection were analyzed. Temporal and compartmental variability of viral populations were quantified using high throughput sequencing and population genetics approaches. HCMV populations were generally stable over time, with ~88% of SNPs displaying similar frequencies. However, samples collected from plasma and urine of the same patient at the same time were highly differentiated with approximately 1700 consensus sequence SNPs (1.2% of the genome) identified between compartments. This inter-compartment differentiation was comparable to the differentiation observed in unrelated hosts. Models of demography (i.e., changes in population size and structure) and positive selection were evaluated to explain the observed patterns of variation. Evidence for strong bottlenecks (>90% reduction in viral population size) was consistent among all patients. From the timing of the bottlenecks, we conclude that fetal infection occurred between 13-18 weeks gestational age in patients analyzed, while colonization of the urine compartment followed roughly 2 months later. The timing of these bottlenecks is consistent with the clinical histories of congenital HCMV infections. We next inferred that positive selection plays a small but measurable role in viral evolution within a single compartment. However, positive selection appears to be a strong and pervasive driver of evolution associated with compartmentalization, affecting ≥ 34 of the 167 open reading frames (~20%) of the genome. This work offers the most detailed map of HCMV in vivo evolution to date and provides evidence that viral populations can be stable or rapidly differentiate, depending on host environment. The application of population genetic methods to these data provides clinically useful information, such as the timing of infection and compartment colonization.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

