The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups
- PMID: 24088323
- PMCID: PMC3850935
- DOI: 10.1186/1471-2229-13-148
The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups
Abstract
Background: WRKY genes encode one of the most abundant groups of transcription factors in higher plants, and its members regulate important biological process such as growth, development, and responses to biotic and abiotic stresses. Although the soybean genome sequence has been published, functional studies on soybean genes still lag behind those of other species.
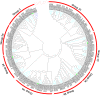
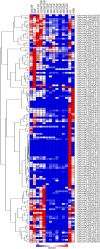
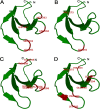
Results: We identified a total of 133 WRKY members in the soybean genome. According to structural features of their encoded proteins and to the phylogenetic tree, the soybean WRKY family could be classified into three groups (groups I, II, and III). A majority of WRKY genes (76.7%; 102 of 133) were segmentally duplicated and 13.5% (18 of 133) of the genes were tandemly duplicated. This pattern was not apparent in Arabidopsis or rice. The transcriptome atlas revealed notable differential expression in either transcript abundance or in expression patterns under normal growth conditions, which indicated wide functional divergence in this family. Furthermore, some critical amino acids were detected using DIVERGE v2.0 in specific comparisons, suggesting that these sites have contributed to functional divergence among groups or subgroups. In addition, site model and branch-site model analyses of positive Darwinian selection (PDS) showed that different selection regimes could have affected the evolution of these groups. Sites with high probabilities of having been under PDS were found in groups I, II c, II e, and III. Together, these results contribute to a detailed understanding of the molecular evolution of the WRKY gene family in soybean.
Conclusions: In this work, all the WRKY genes, which were generated mainly through segmental duplication, were identified in the soybean genome. Moreover, differential expression and functional divergence of the duplicated WRKY genes were two major features of this family throughout their evolutionary history. Positive selection analysis revealed that the different groups have different evolutionary rates. Together, these results contribute to a detailed understanding of the molecular evolution of the WRKY gene family in soybean.
Figures


References
-
- Ishiguro N. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol Gen Genet. 1994;244:563–571. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

