Characterization of GM events by insert knowledge adapted re-sequencing approaches
- PMID: 24088728
- PMCID: PMC3789143
- DOI: 10.1038/srep02839
Characterization of GM events by insert knowledge adapted re-sequencing approaches
Abstract
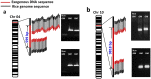
Detection methods and data from molecular characterization of genetically modified (GM) events are needed by stakeholders of public risk assessors and regulators. Generally, the molecular characteristics of GM events are incomprehensively revealed by current approaches and biased towards detecting transformation vector derived sequences. GM events are classified based on available knowledge of the sequences of vectors and inserts (insert knowledge). Herein we present three insert knowledge-adapted approaches for characterization GM events (TT51-1 and T1c-19 rice as examples) based on paired-end re-sequencing with the advantages of comprehensiveness, accuracy, and automation. The comprehensive molecular characteristics of two rice events were revealed with additional unintended insertions comparing with the results from PCR and Southern blotting. Comprehensive transgene characterization of TT51-1 and T1c-19 is shown to be independent of a priori knowledge of the insert and vector sequences employing the developed approaches. This provides an opportunity to identify and characterize also unknown GM events.
Figures


References
-
- Secretariat of the Convention on Biological Diversity. Secretariat of the Convention of Biological Diversity (2000).
-
- Codex Alimentarius. Guideline for the conduct of food safety assessment of foods derived from recombinant-DNA plants. CAC/GL 45, 1–18 (2003).
-
- Sparrow P. GM risk assessment. Mol biotechnol 44, 267–275 (2010). - PubMed
-
- Miraglia M. et al. Detection and traceability of genetically modified organisms in the food production chain. Food Chem Toxicol 42, 1157–1180 (2004). - PubMed
-
- Padgette S. R. et al. Development, identification, and characterization of a glyphosate-tolerant soybean line. Crop Sci 35, 1451–1461 (1995).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

