Functional interaction between autophagy and ciliogenesis
- PMID: 24089209
- PMCID: PMC3896125
- DOI: 10.1038/nature12639
Functional interaction between autophagy and ciliogenesis
Abstract
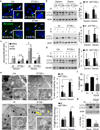
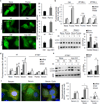
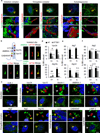
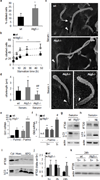
Nutrient deprivation is a stimulus shared by both autophagy and the formation of primary cilia. The recently discovered role of primary cilia in nutrient sensing and signalling motivated us to explore the possible functional interactions between this signalling hub and autophagy. Here we show that part of the molecular machinery involved in ciliogenesis also participates in the early steps of the autophagic process. Signalling from the cilia, such as that from the Hedgehog pathway, induces autophagy by acting directly on essential autophagy-related proteins strategically located in the base of the cilium by ciliary trafficking proteins. Whereas abrogation of ciliogenesis partially inhibits autophagy, blockage of autophagy enhances primary cilia growth and cilia-associated signalling during normal nutritional conditions. We propose that basal autophagy regulates ciliary growth through the degradation of proteins required for intraflagellar transport. Compromised ability to activate the autophagic response may underlie some common ciliopathies.
Figures

Comment in
-
Cell biology: Molecular clearance at the cell's antenna.Nature. 2013 Oct 10;502(7470):180-1. doi: 10.1038/nature12693. Epub 2013 Oct 2. Nature. 2013. PMID: 24089206 No abstract available.
-
Cytoskeleton: Autophagy and ciliogenesis come together.Nat Rev Mol Cell Biol. 2013 Nov;14(11):687. doi: 10.1038/nrm3686. Epub 2013 Oct 16. Nat Rev Mol Cell Biol. 2013. PMID: 24129416 No abstract available.
-
Cilia grow by taking a bite out of the cell.Dev Cell. 2013 Oct 28;27(2):126-127. doi: 10.1016/j.devcel.2013.10.013. Dev Cell. 2013. PMID: 24176638 Free PMC article.
References
-
- Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol. 2013;25:455–460. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

