Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes
- PMID: 24089468
- PMCID: PMC4007716
- DOI: 10.1242/dev.099390
Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes
Abstract
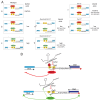
Several lines of evidence suggest that long non-coding RNA (lncRNA)-dependent mechanisms regulate transcription and CpG DNA methylation. Whereas CpG island methylation has been studied in detail, the significance of enhancer DNA methylation and its relationship with lncRNAs is relatively unexplored. Previous experiments proposed that the ultraconserved lncRNA Evf2 represses transcription through Dlx6 antisense (Dlx6as) transcription and methyl-CpG binding protein (MECP2) recruitment to the Dlx5/6 ultraconserved DNA regulatory enhancer (Dlx5/6ei) in embryonic day 13.5 medial ganglionic eminence (E13.5 MGE). Here, genetic epistasis experiments show that MECP2 transcriptional repression of Evf2 and Dlx5, but not Dlx6, occurs through antagonism of DLX1/2 in E13.5 MGE. Analysis of E13.5 MGE from mice lacking Evf2 and of partially rescued Evf2 transgenic mice shows that Evf2 prevents site-specific CpG DNA methylation of Dlx5/6ei in trans, without altering Dlx5/6 expression. Dlx1/2 loss increases CpG DNA methylation, whereas Mecp2 loss does not affect Dlx5/6ei methylation. Based on these studies, we propose a model in which Evf2 inhibits enhancer DNA methylation, effectively modulating competition between the DLX1/2 activator and MECP2 repressor. Evf2 antisense transcription and Evf2-dependent balanced recruitment of activator and repressor proteins enables differential transcriptional control of adjacent genes with shared DNA regulatory elements.
Keywords: Forebrain; MECP2; Mouse; Ultraconserved enhancer methylation.
Figures
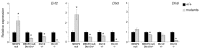




References
-
- Anderson S. A., Eisenstat D. D., Shi L., Rubenstein J. L. (1997a). Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474–476 - PubMed
-
- Anderson S. A., Qiu M., Bulfone A., Eisenstat D. D., Meneses J., Pedersen R., Rubenstein J. L. (1997b). Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19, 27–37 - PubMed
-
- Barlow D. P. (2011). Genomic imprinting: a mammalian epigenetic discovery model. Annu. Rev. Genet. 45, 379–403 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

