Graded encoding of food odor value in the Drosophila brain
- PMID: 24089477
- PMCID: PMC3787495
- DOI: 10.1523/JNEUROSCI.2605-13.2013
Graded encoding of food odor value in the Drosophila brain
Abstract
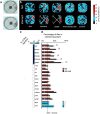
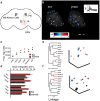
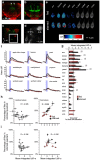
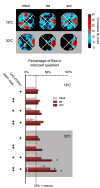
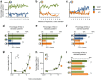
Odors are highly evocative, yet how and where in the brain odors derive meaning remains unknown. Our analysis of the Drosophila brain extends the role of a small number of hunger-sensing neurons to include food-odor value representation. In vivo two-photon calcium imaging shows the amplitude of food odor-evoked activity in neurons expressing Drosophila neuropeptide F (dNPF), the neuropeptide Y homolog, strongly correlates with food-odor attractiveness. Hunger elevates neural and behavioral responses to food odors only, although food odors that elicit attraction in the fed state also evoke heightened dNPF activity in fed flies. Inactivation of a subset of dNPF-expressing neurons or silencing dNPF receptors abolishes food-odor attractiveness, whereas genetically enhanced dNPF activity not only increases food-odor attractiveness but promotes attraction to aversive odors. Varying the amount of presented odor produces matching graded neural and behavioral curves, which can function to predict preference between odors. We thus demonstrate a possible motivationally scaled neural "value signal" accessible from uniquely identifiable cells.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
