Evasion of superinfection exclusion and elimination of primary viral RNA by an adapted strain of hepatitis C virus
- PMID: 24089557
- PMCID: PMC3838274
- DOI: 10.1128/JVI.02465-13
Evasion of superinfection exclusion and elimination of primary viral RNA by an adapted strain of hepatitis C virus
Abstract
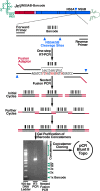
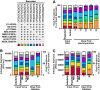
Cells that are productively infected by hepatitis C virus (HCV) are refractory to a second infection by HCV via a block in viral replication known as superinfection exclusion. The block occurs at a postentry step and likely involves translation or replication of the secondary viral RNA, but the mechanism is largely unknown. To characterize HCV superinfection exclusion, we selected for an HCV variant that could overcome the block. We produced a high-titer HC-J6/JFH1 (Jc1) viral genome with a fluorescent reporter inserted between NS5A and NS5B and used it to infect Huh7.5 cells containing a Jc1 replicon. With multiple passages of these infected cells, we isolated an HCV variant that can superinfect cells at high levels. Notably, the superinfectious virus rapidly cleared the primary replicon from superinfected cells. Viral competition experiments, using a novel strategy of sequence-barcoding viral strains, as well as superinfection of replicon cells demonstrated that mutations in E1, p7, NS5A, and the poly(U/UC) tract of the 3' untranslated region were important for superinfection. Furthermore, these mutations dramatically increased the infectivity of the virus in naive cells. Interestingly, viruses with a shorter poly(U/UC) and an NS5A domain II mutation were most effective in overcoming the postentry block. Neither of these changes affected viral RNA translation, indicating that the major barrier to postentry exclusion occurs at viral RNA replication. The evolution of the ability to superinfect after less than a month in culture and the concomitant exclusion of the primary replicon suggest that superinfection exclusion dramatically affects viral fitness and dynamics in vivo.
Figures


Similar articles
-
Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells.J Virol. 2002 Mar;76(6):2997-3006. doi: 10.1128/jvi.76.6.2997-3006.2002. J Virol. 2002. PMID: 11861865 Free PMC article.
-
Adaptive Mutations Enhance Assembly and Cell-to-Cell Transmission of a High-Titer Hepatitis C Virus Genotype 5a Core-NS2 JFH1-Based Recombinant.J Virol. 2015 Aug;89(15):7758-75. doi: 10.1128/JVI.00039-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995244 Free PMC article.
-
Cell culture-adaptive mutations in hepatitis C virus promote viral production by enhancing viral replication and release.World J Gastroenterol. 2018 Mar 28;24(12):1299-1311. doi: 10.3748/wjg.v24.i12.1299. World J Gastroenterol. 2018. PMID: 29599605 Free PMC article.
-
[Role of interferon-sensitivity-determining region in replication of hepatitis C virus].Nihon Rinsho. 2004 Jul;62 Suppl 7(Pt 1):89-95. Nihon Rinsho. 2004. PMID: 15359770 Review. Japanese. No abstract available.
-
Hepatitis C virus RNA replication.Curr Top Microbiol Immunol. 2013;369:167-98. doi: 10.1007/978-3-642-27340-7_7. Curr Top Microbiol Immunol. 2013. PMID: 23463201 Free PMC article. Review.
Cited by
-
Lipid Droplet Isolation for Quantitative Mass Spectrometry Analysis.J Vis Exp. 2017 Apr 17;(122):55585. doi: 10.3791/55585. J Vis Exp. 2017. PMID: 28448054 Free PMC article.
-
Classical Swine Fever Virus vs. Classical Swine Fever Virus: The Superinfection Exclusion Phenomenon in Experimentally Infected Wild Boar.PLoS One. 2016 Feb 26;11(2):e0149469. doi: 10.1371/journal.pone.0149469. eCollection 2016. PLoS One. 2016. PMID: 26919741 Free PMC article.
-
A viral protein mediates superinfection exclusion at the whole-organism level but is not required for exclusion at the cellular level.J Virol. 2014 Oct;88(19):11327-38. doi: 10.1128/JVI.01612-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031351 Free PMC article.
-
Mimicking superinfection exclusion disrupts alphavirus infection and transmission in the yellow fever mosquito Aedes aegypti.Proc Natl Acad Sci U S A. 2023 Sep 12;120(37):e2303080120. doi: 10.1073/pnas.2303080120. Epub 2023 Sep 5. Proc Natl Acad Sci U S A. 2023. PMID: 37669371 Free PMC article.
-
Clonality and intracellular polyploidy in virus evolution and pathogenesis.Proc Natl Acad Sci U S A. 2015 Jul 21;112(29):8887-92. doi: 10.1073/pnas.1501715112. Epub 2015 Jul 20. Proc Natl Acad Sci U S A. 2015. PMID: 26195777 Free PMC article.
References
-
- Michel N, Allespach I, Venzke S, Fackler OT, Keppler OT. 2005. The Nef protein of human immunodeficiency virus establishes superinfection immunity by a dual strategy to downregulate cell surface CCR5 and CD4. Curr. Biol. 15:714–723 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

