A quantitative measure for protein conformational heterogeneity
- PMID: 24089719
- PMCID: PMC3724800
- DOI: 10.1063/1.4812791
A quantitative measure for protein conformational heterogeneity
Abstract
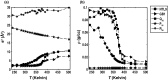
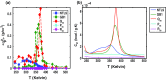
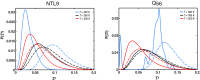
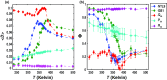
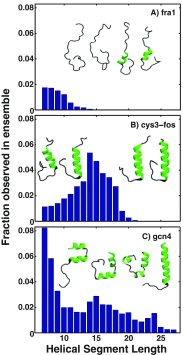
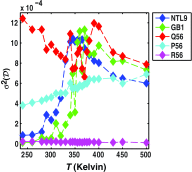
Conformational heterogeneity is a defining characteristic of proteins. Intrinsically disordered proteins (IDPs) and denatured state ensembles are extreme manifestations of this heterogeneity. Inferences regarding globule versus coil formation can be drawn from analysis of polymeric properties such as average size, shape, and density fluctuations. Here we introduce a new parameter to quantify the degree of conformational heterogeneity within an ensemble to complement polymeric descriptors. The design of this parameter is guided by the need to distinguish between systems that couple their unfolding-folding transitions with coil-to-globule transitions and those systems that undergo coil-to-globule transitions with no evidence of acquiring a homogeneous ensemble of conformations upon collapse. The approach is as follows: Each conformation in an ensemble is converted into a conformational vector where the elements are inter-residue distances. Similarity between pairs of conformations is quantified using the projection between the corresponding conformational vectors. An ensemble of conformations yields a distribution of pairwise projections, which is converted into a distribution of pairwise conformational dissimilarities. The first moment of this dissimilarity distribution is normalized against the first moment of the distribution obtained by comparing conformations from the ensemble of interest to conformations drawn from a Flory random coil model. The latter sets an upper bound on conformational heterogeneity thus ensuring that the proposed measure for intra-ensemble heterogeneity is properly calibrated and can be used to compare ensembles for different sequences and across different temperatures. The new measure of conformational heterogeneity will be useful in quantitative studies of coupled folding and binding of IDPs and in de novo sequence design efforts that are geared toward controlling the degree of heterogeneity in unbound forms of IDPs.
Figures

References
-
- Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausio J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., Bailey R. W., Griswold M. D., Chiu W., Garner E. C., and Obradovic Z., J. Mol. Graphics Modell. 19, 26 (2001). 10.1016/S1093-3263(00)00138-8 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

