Identification of novel small molecule inhibitors of centrosome clustering in cancer cells
- PMID: 24091544
- PMCID: PMC3858562
- DOI: 10.18632/oncotarget.1198
Identification of novel small molecule inhibitors of centrosome clustering in cancer cells
Abstract
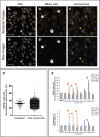
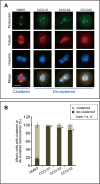
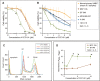
Most normal cells have two centrosomes that form bipolar spindles during mitosis, while cancer cells often contain more than two, or "supernumerary" centrosomes. Such cancer cells achieve bipolar division by clustering their centrosomes into two functional poles, and inhibiting this process then leads to cancer-specific cell death. A major problem with clinically used anti-mitotic drugs, such as paclitaxel, is their toxicity in normal cells. To discover new compounds with greater specificity for cancer cells, we established a high-content screen for agents that block centrosome clustering in BT-549 cells, a breast cancer cell line that harbors supernumerary centrosomes. Using this screen, we identified 14 compounds that inhibit centrosome clustering and induce mitotic arrest. Some of these compounds were structurally similar, suggesting a common structural motif important for preventing centrosome clustering. We next compared the effects of these compounds on the growth of several breast and other cancer cell lines, an immortalized normal human mammary epithelial cell line, and progenitor-enriched primary normal human mammary epithelial cells. From these comparisons, we found some compounds that kill breast cancer cells, but not their normal epithelial counterparts, suggesting their potential for targeted therapy. One of these compounds, N2-(3-pyridylmethyl)-5-nitro-2-furamide (Centrosome Clustering Chemical Inhibitor-01, CCCI-01), that showed the greatest differential response in this screen was confirmed to have selective effects on cancer as compared to normal breast progenitors using more precise apoptosis induction and clonogenic growth endpoints. The concentration of CCCI-01 that killed cancer cells in the clonogenic assay spared normal human bone marrow hematopoietic progenitors in the colony-forming cell assay, indicating a potential therapeutic window for CCCI-01, whose selectivity might be further improved by optimizing the compound. Immunofluorescence analysis showed that treatment with CCCI-01 lead to multipolar spindles in BT-549, while maintaining bipolar spindles in the normal primary human mammary epithelial cells. Since centrosome clustering is a complex process involving multiple pathways, the 14 compounds identified in this study provide a potentially novel means to developing non-cross-resistant anti-cancer drugs that block centrosome clustering.
Figures




References
-
- Zyss D, Gergely F. Centrosome function in cancer: guilty or innocent? Trends Cell Biol. 2009;19(7):334–346. - PubMed
-
- Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nature Reviews Cancer. 2007;7(12):911–924. - PubMed
-
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307(5706):127–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

