Personalized prediction of EGFR mutation-induced drug resistance in lung cancer
- PMID: 24092472
- PMCID: PMC3790204
- DOI: 10.1038/srep02855
Personalized prediction of EGFR mutation-induced drug resistance in lung cancer
Abstract
EGFR mutation-induced drug resistance has significantly impaired the potency of small molecule tyrosine kinase inhibitors in lung cancer treatment. Computational approaches can provide powerful and efficient techniques in the investigation of drug resistance. In our work, the EGFR mutation feature is characterized by the energy components of binding free energy (concerning the mutant-inhibitor complex), and we combine it with specific personal features for 168 clinical subjects to construct a personalized drug resistance prediction model. The 3D structure of an EGFR mutant is computationally predicted from its protein sequence, after which the dynamics of the bound mutant-inhibitor complex is simulated via AMBER and the binding free energy of the complex is calculated based on the dynamics. The utilization of extreme learning machines and leave-one-out cross-validation promises a successful identification of resistant subjects with high accuracy. Overall, our study demonstrates advantages in the development of personalized medicine/therapy design and innovative drug discovery.
Figures

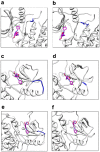
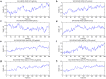


References
-
- Lynch T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350, 2129–2139 (2004). - PubMed
-
- Kobayashi S. et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. New Engl J Med 352, 786–792 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

