Integrative annotation of variants from 1092 humans: application to cancer genomics
- PMID: 24092746
- PMCID: PMC3947637
- DOI: 10.1126/science.1235587
Integrative annotation of variants from 1092 humans: application to cancer genomics
Abstract
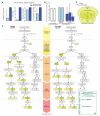
Interpreting variants, especially noncoding ones, in the increasing number of personal genomes is challenging. We used patterns of polymorphisms in functionally annotated regions in 1092 humans to identify deleterious variants; then we experimentally validated candidates. We analyzed both coding and noncoding regions, with the former corroborating the latter. We found regions particularly sensitive to mutations ("ultrasensitive") and variants that are disruptive because of mechanistic effects on transcription-factor binding (that is, "motif-breakers"). We also found variants in regions with higher network centrality tend to be deleterious. Insertions and deletions followed a similar pattern to single-nucleotide variants, with some notable exceptions (e.g., certain deletions and enhancers). On the basis of these patterns, we developed a computational tool (FunSeq), whose application to ~90 cancer genomes reveals nearly a hundred candidate noncoding drivers.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG002898/HG/NHGRI NIH HHS/United States
- R01 CA152057/CA/NCI NIH HHS/United States
- U01 HG005718/HG/NHGRI NIH HHS/United States
- R01 HG004719/HG/NHGRI NIH HHS/United States
- WT090532/WT_/Wellcome Trust/United Kingdom
- WT095908/WT_/Wellcome Trust/United Kingdom
- WT098051/WT_/Wellcome Trust/United Kingdom
- CA167824/CA/NCI NIH HHS/United States
- R01CA152057/CA/NCI NIH HHS/United States
- R01 GM097358/GM/NIGMS NIH HHS/United States
- 085532/WT_/Wellcome Trust/United Kingdom
- U01HG6513/HG/NHGRI NIH HHS/United States
- WT085532/WT_/Wellcome Trust/United Kingdom
- HG005718/HG/NHGRI NIH HHS/United States
- U01 HG006513/HG/NHGRI NIH HHS/United States
- R01HG4719/HG/NHGRI NIH HHS/United States
- 098051/WT_/Wellcome Trust/United Kingdom
- G12 MD007579/MD/NIMHD NIH HHS/United States
- P20 MD006899/MD/NIMHD NIH HHS/United States
- R01 CA167824/CA/NCI NIH HHS/United States
- 090532/WT_/Wellcome Trust/United Kingdom
- 095908/WT_/Wellcome Trust/United Kingdom
- R01 CA166661/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- GM104424/GM/NIGMS NIH HHS/United States
- U41 HG007000/HG/NHGRI NIH HHS/United States
- UL1 TR000457/TR/NCATS NIH HHS/United States
- HG007000/HG/NHGRI NIH HHS/United States
- U01 CA111275/CA/NCI NIH HHS/United States
- R01 GM104424/GM/NIGMS NIH HHS/United States
- G12 RR003050/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

