Filovirus replication and transcription
- PMID: 24093048
- PMCID: PMC3787895
- DOI: 10.2217/17460794.2.2.205
Filovirus replication and transcription
Abstract
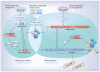
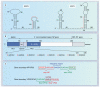
The highly pathogenic filoviruses, Marburg and Ebola virus, belong to the nonsegmented negative-sense RNA viruses of the order Mononegavirales. The mode of replication and transcription is similar for these viruses. On one hand, the negative-sense RNA genome serves as a template for replication, to generate progeny genomes, and, on the other hand, for transcription, to produce mRNAs. Despite the similarities in the replication/transcription strategy, filoviruses have evolved structural and functional properties that are unique among the nonsegmented negative-sense RNA viruses. Moreover, there are also striking differences in the replication and transcription mechanisms of Marburg and Ebola virus. This includes nucleocapsid formation, the structure of the genomic replication promoter, the protein requirement for transcription and the use of mRNA editing. In this article, the current knowledge of the replication and transcription strategy of Marburg and Ebola virus is reviewed, with focus on the observed differences.
Keywords: ebola virus; hemorrhagic fever; marburg virus; nonsegmented negative-sense RNA viruses; nucleocapsid complex; replication; reverse genetics; transcription.
Figures


References
-
- Hensley LE, Jones SM, Feldmann H, Jahrling PB, Geisbert TW. Ebola and Marburg viruses: pathogenesis and development of countermeasures. Curr. Mol. Med. 2005;5:761–772. - PubMed
-
- Feldmann H, Geisbert TW, Jahrling PB, et al. In: Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Elsevier/Academic Press; San Diego, USA: 2005. pp. 645–653.
-
- Wang L, Harcourt BH, Yu M, et al. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 2001;3:279–287. - PubMed
-
- Sanchez A, Rollin PE. Complete genome sequence of an Ebola virus (Sudan species) responsible for a 2000 outbreak of human disease in Uganda. Virus Res. 2005;113:16–25. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
