Quantitative DNA methylation analyses reveal stage dependent DNA methylation and association to clinico-pathological factors in breast tumors
- PMID: 24093668
- PMCID: PMC3819713
- DOI: 10.1186/1471-2407-13-456
Quantitative DNA methylation analyses reveal stage dependent DNA methylation and association to clinico-pathological factors in breast tumors
Abstract
Background: Aberrant DNA methylation of regulatory genes has frequently been found in human breast cancers and correlated to clinical outcome. In the present study we investigate stage specific changes in the DNA methylation patterns in order to identify valuable markers to understand how these changes affect breast cancer progression.
Methods: Quantitative DNA methylation analyses of 12 candidate genes ABCB1, BRCCA1, CDKN2A, ESR1, GSTP1, IGF2, MGMT, HMLH1, PPP2R2B, PTEN, RASSF1A and FOXC1 was performed by pyrosequencing a series of 238 breast cancer tissue samples from DCIS to invasive tumors stage I to IV.
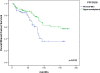
Results: Significant differences in methylation levels between the DCIS and invasive stage II tumors were observed for six genes RASSF1A, CDKN2A, MGMT, ABCB1, GSTP1 and FOXC1. RASSF1A, ABCB1 and GSTP1 showed significantly higher methylation levels in late stage compared to the early stage breast carcinoma. Z-score analysis revealed significantly lower methylation levels in DCIS and stage I tumors compared with stage II, III and IV tumors. Methylation levels of PTEN, PPP2R2B, FOXC1, ABCB1 and BRCA1 were lower in tumors harboring TP53 mutations then in tumors with wild type TP53. Z-score analysis showed that TP53 mutated tumors had significantly lower overall methylation levels compared to tumors with wild type TP53. Methylation levels of RASSF1A, PPP2R2B, GSTP1 and FOXC1 were higher in ER positive vs. ER negative tumors and methylation levels of PTEN and CDKN2A were higher in HER2 positive vs. HER2 negative tumors. Z-score analysis also showed that HER2 positive tumors had significantly higher z-scores of methylation compared to the HER2 negative tumors. Univariate survival analysis identifies methylation status of PPP2R2B as significant predictor of overall survival and breast cancer specific survival.
Conclusions: In the present study we report that the level of aberrant DNA methylation is higher in late stage compared with early stage of invasive breast cancers and DCIS for genes mentioned above.
Figures



Similar articles
-
Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer.Breast Cancer Res. 2010;12(1):R3. doi: 10.1186/bcr2466. Epub 2010 Jan 7. Breast Cancer Res. 2010. PMID: 20056007 Free PMC article.
-
DNA methylation profiling in doxorubicin treated primary locally advanced breast tumours identifies novel genes associated with survival and treatment response.Mol Cancer. 2010 Mar 25;9:68. doi: 10.1186/1476-4598-9-68. Mol Cancer. 2010. PMID: 20338046 Free PMC article.
-
Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors.Breast Cancer Res. 2008;10(3):R46. doi: 10.1186/bcr2098. Epub 2008 May 16. Breast Cancer Res. 2008. PMID: 18485221 Free PMC article.
-
p53 in breast cancer subtypes and new insights into response to chemotherapy.Breast. 2013 Aug;22 Suppl 2:S27-9. doi: 10.1016/j.breast.2013.07.005. Breast. 2013. PMID: 24074787 Review.
-
Novel insights into DNA methylation-based epigenetic regulation of breast tumor angiogenesis.Int Rev Cell Mol Biol. 2023;380:63-96. doi: 10.1016/bs.ircmb.2023.04.002. Epub 2023 May 18. Int Rev Cell Mol Biol. 2023. PMID: 37657860 Review.
Cited by
-
Active and secondhand smoke exposure throughout life and DNA methylation in breast tumors.Cancer Causes Control. 2019 Jan;30(1):53-62. doi: 10.1007/s10552-018-1102-4. Epub 2019 Jan 7. Cancer Causes Control. 2019. PMID: 30617699 Free PMC article.
-
Heterogeneous DNA Methylation Patterns in the GSTP1 Promoter Lead to Discordant Results between Assay Technologies and Impede Its Implementation as Epigenetic Biomarkers in Breast Cancer.Genes (Basel). 2015 Sep 17;6(3):878-900. doi: 10.3390/genes6030878. Genes (Basel). 2015. PMID: 26393654 Free PMC article.
-
Validation of DNA promoter hypermethylation biomarkers in breast cancer--a short report.Cell Oncol (Dordr). 2014 Aug;37(4):297-303. doi: 10.1007/s13402-014-0189-1. Epub 2014 Aug 16. Cell Oncol (Dordr). 2014. PMID: 25123395
-
FOXC1: an emerging marker and therapeutic target for cancer.Oncogene. 2017 Jul 13;36(28):3957-3963. doi: 10.1038/onc.2017.48. Epub 2017 Mar 13. Oncogene. 2017. PMID: 28288141 Free PMC article. Review.
-
The forkhead box C1 (FOXC1) transcription factor is downregulated in acute promyelocytic leukemia.Oncotarget. 2017 Sep 20;8(48):84074-84085. doi: 10.18632/oncotarget.21101. eCollection 2017 Oct 13. Oncotarget. 2017. PMID: 29137406 Free PMC article.
References
-
- Dejeux E, Rønneberg JA, Solvang H, Bukholm I, Geisler S, Aas T, Gut IG, Børresen-Dale AL, Lønning PE, Kristensen VN, Tost J. DNA methylation profiling in doxorubicin treated primary locally advanced breast tumours identifies novel genes associated with survival and treatment response. Mol Cancer. 2010;9:68. doi: 10.1186/1476-4598-9-68. - DOI - PMC - PubMed
-
- Muggerud AA, Rønneberg JA, Wärnberg F, Botling J, Busato F, Jovanovic J, Solvang H, Bukholm I, Børresen-Dale AL, Kristensen VN, Sørlie T, Tost J. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res. 2010;12:R3. doi: 10.1186/bcr2466. - DOI - PMC - PubMed
-
- Geisler S, Børresen-Dale AL, Johnsen H, Aas T, Geisler J, Akslen LA, Anker G, Lønning PE. TP53 gene mutations predict the response to neoadjuvant treatment with 5-fluorouracil and mitomycin in locally advanced breast cancer. Clin Cancer Res. 2003;9:5582–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

