"A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids"
- PMID: 24094114
- PMCID: PMC3852598
- DOI: 10.1186/1471-2164-14-683
"A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids"
Abstract
Background: Modern banana cultivars are primarily interspecific triploid hybrids of two species, Musa acuminata and Musa balbisiana, which respectively contribute the A- and B-genomes. The M. balbisiana genome has been associated with improved vigour and tolerance to biotic and abiotic stresses and is thus a target for Musa breeding programs. However, while a reference M. acuminata genome has recently been released (Nature 488:213-217, 2012), little sequence data is available for the corresponding B-genome.To address these problems we carried out Next Generation gDNA sequencing of the wild diploid M. balbisiana variety 'Pisang Klutuk Wulung' (PKW). Our strategy was to align PKW gDNA reads against the published A-genome and to extract the mapped consensus sequences for subsequent rounds of evaluation and gene annotation.
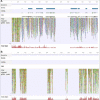
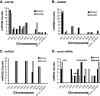
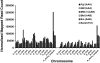
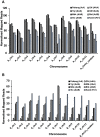
Results: The resulting B-genome is 79% the size of the A-genome, and contains 36,638 predicted functional gene sequences which is nearly identical to the 36,542 of the A-genome. There is substantial sequence divergence from the A-genome at a frequency of 1 homozygous SNP per 23.1 bp, and a high degree of heterozygosity corresponding to one heterozygous SNP per 55.9 bp. Using expressed small RNA data, a similar number of microRNA sequences were predicted in both A- and B-genomes, but additional novel miRNAs were detected, including some that are unique to each genome. The usefulness of this B-genome sequence was evaluated by mapping RNA-seq data from a set of triploid AAA and AAB hybrids simultaneously to both genomes. Results for the plantains demonstrated the expected 2:1 distribution of reads across the A- and B-genomes, but for the AAA genomes, results show they contain regions of significant homology to the B-genome supporting proposals that there has been a history of interspecific recombination between homeologous A and B chromosomes in Musa hybrids.
Conclusions: We have generated and annotated a draft reference Musa B-genome and demonstrate that this can be used for molecular genetic mapping of gene transcripts and small RNA expression data from several allopolyploid banana cultivars. This draft therefore represents a valuable resource to support the study of metabolism in inter- and intraspecific triploid Musa hybrids and to help direct breeding programs.
Figures

References
-
- De LE, Vrydaghs L, De Maret P, Xavier P, Denham T. Why bananas matter: an introduction to the history of banana domestication. Ethnobot Res Appl. 2009;7:165–177.
-
- FAOSTAT, banana consumption. http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567#ancor.
-
- FAOSTAT, banana exports. http://faostat.fao.org/site/535/DesktopDefault.aspx?PageID=535#ancor.
-
- Raboin L, Carreel F, Noyer J. Diploid ancestors of triploid export banana cultivars: molecular identification of 2n restitution gamete donors and n gamete donors. Mol Breed. 2005;16:333–341. doi: 10.1007/s11032-005-2452-7. - DOI
-
- Boonruangrod R, Fluch ÆS, Burg ÆK. Elucidation of origin of the present day hybrid banana cultivars using the 5’ ETS rDNA sequence information. Mol Breed. 2009;24:77–91. doi: 10.1007/s11032-009-9273-z. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

