Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors
- PMID: 24094326
- PMCID: PMC3888026
- DOI: 10.1016/j.stem.2013.09.002
Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors
Erratum in
-
Induction of Multipotential Hematopoietic Progenitors from Human Pluripotent Stem Cells via Respecification of Lineage-Restricted Precursors.Cell Stem Cell. 2025 Jun 5;32(6):1025. doi: 10.1016/j.stem.2025.04.009. Epub 2025 Apr 29. Cell Stem Cell. 2025. PMID: 40306277 Free PMC article. No abstract available.
Abstract
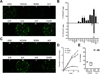
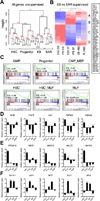
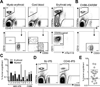
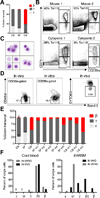
Human pluripotent stem cells (hPSCs) represent a promising source of patient-specific cells for disease modeling, drug screens, and cellular therapies. However, the inability to derive engraftable human hematopoietic stem and progenitor cells (HSPCs) has limited their characterization to in vitro assays. We report a strategy to respecify lineage-restricted CD34(+)CD45(+) myeloid precursors derived from hPSCs into multilineage progenitors that can be expanded in vitro and engrafted in vivo. HOXA9, ERG, and RORA conferred self-renewal and multilineage potential in vitro and maintained primitive CD34(+)CD38(-) cells. Screening cells via transplantation revealed that two additional factors, SOX4 and MYB, conferred engraftment. Progenitors specified with all five factors gave rise to reproducible short-term engraftment with myeloid and erythroid lineages. Erythroid precursors underwent hemoglobin switching in vivo, silencing embryonic and activating adult globin expression. Our combinatorial screening approach establishes a strategy for obtaining transcription-factor-mediated engraftment of blood progenitors from human pluripotent cells.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



References
-
- Amsellem S, Pflumio F, Bardinet D, Izac B, Charneau P, Romeo PH, Dubart-Kupperschmitt A, Fichelson S. Ex vivo expansion of human hematopoietic stem cells by direct delivery of the HOXB4 homeoprotein. Nature medicine. 2003;9:1423–1427. - PubMed
-
- Anstee DJ, Gampel A, Toye AM. Ex-vivo generation of human red cells for transfusion. Current opinion in hematology. 2012;19:163–169. - PubMed
-
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

