Nuclear repulsion enables division autonomy in a single cytoplasm
- PMID: 24094857
- PMCID: PMC4085259
- DOI: 10.1016/j.cub.2013.07.076
Nuclear repulsion enables division autonomy in a single cytoplasm
Abstract
Background: Current models of cell-cycle control, based on classic studies of fused cells, predict that nuclei in a shared cytoplasm respond to the same CDK activities to undergo synchronous cycling. However, synchrony is rarely observed in naturally occurring syncytia, such as the multinucleate fungus Ashbya gossypii. In this system, nuclei divide asynchronously, raising the question of how nuclear timing differences are maintained despite sharing a common milieu.
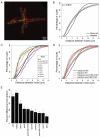
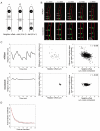
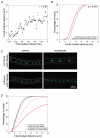
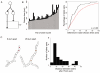
Results: We observe that neighboring nuclei are highly variable in division-cycle duration and that neighbors repel one another to space apart and demarcate their own cytoplasmic territories. The size of these territories increases as a nucleus approaches mitosis and can influence cycling rates. This nonrandom nuclear spacing is regulated by microtubules and is required for nuclear asynchrony, as nuclei that transiently come in very close proximity will partially synchronize. Sister nuclei born of the same mitosis are generally not persistent neighbors over their lifetimes yet remarkably retain similar division cycle times. This indicates that nuclei carry a memory of their birth state that influences their division timing and supports that nuclei subdivide a common cytosol into functionally distinct yet mobile compartments.
Conclusions: These findings support that nuclei use cytoplasmic microtubules to establish "cells within cells." Individual compartments appear to push against one another to compete for cytoplasmic territory and insulate the division cycle. This provides a mechanism by which syncytial nuclei can spatially organize cell-cycle signaling and suggests size control can act in a system without physical boundaries.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


References
-
- Rao PN, Johnson RT. Premature chromosome condensation: a mechanism for the elimination of chromosomes in virus-fused cells. J. Cell. Sci. 1972;10:495–513. - PubMed
-
- Johnson RT, Rao PN. Nucleo-cytoplasmic interactions in the acheivement of nuclear synchrony in DNA synthesis and mitosis in multinucleate cells. Biol Rev Camb Philos Soc. 1971;46:97–155. - PubMed
-
- Rao PN, Johnson RT. Mammalian cell fusion. IV. Regulation of chromosome formation from interphase nuclei by various chemical compounds. J. Cell. Physiol. 1971;78:217–223. - PubMed
-
- Johnson RT, Rao PN. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature. 1970;226:717–722. - PubMed
-
- Johnson RT, Rao PN, Hughes HD. Mammalian cell fusion III. A hela cell inducer of premature chromosome condensation active in cells from a variety of animal species. J. Cell. Physiol. 1970;76:151–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

