Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor
- PMID: 24096423
- PMCID: PMC3837824
- DOI: 10.1128/AEM.02090-13
Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor
Abstract
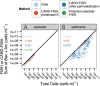
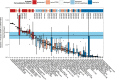
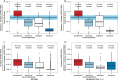
There is no universally accepted method to quantify bacteria and archaea in seawater and marine sediments, and different methods have produced conflicting results with the same samples. To identify best practices, we compiled data from 65 studies, plus our own measurements, in which bacteria and archaea were quantified with fluorescent in situ hybridization (FISH), catalyzed reporter deposition FISH (CARD-FISH), polyribonucleotide FISH, or quantitative PCR (qPCR). To estimate efficiency, we defined "yield" to be the sum of bacteria and archaea counted by these techniques divided by the total number of cells. In seawater, the yield was high (median, 71%) and was similar for FISH, CARD-FISH, and polyribonucleotide FISH. In sediments, only measurements by CARD-FISH in which archaeal cells were permeabilized with proteinase K showed high yields (median, 84%). Therefore, the majority of cells in both environments appear to be alive, since they contain intact ribosomes. In sediments, the sum of bacterial and archaeal 16S rRNA gene qPCR counts was not closely related to cell counts, even after accounting for variations in copy numbers per genome. However, qPCR measurements were precise relative to other qPCR measurements made on the same samples. qPCR is therefore a reliable relative quantification method. Inconsistent results for the relative abundance of bacteria versus archaea in deep subsurface sediments were resolved by the removal of CARD-FISH measurements in which lysozyme was used to permeabilize archaeal cells and qPCR measurements which used ARCH516 as an archaeal primer or TaqMan probe. Data from best-practice methods showed that archaea and bacteria decreased as the depth in seawater and marine sediments increased, although archaea decreased more slowly.
Figures


References
-
- Hoehler TM, Jørgensen BB. 2013. Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11:83–94 - PubMed
-
- Schippers A, Neretin LN, Kallmeyer J, Ferdelman TG, Cragg BA, Parkes RJ, Jørgensen BB. 2005. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 61:861–864 - PubMed
-
- Schippers A, Neretin LN. 2006. Quantification of microbial communities in near-surface and deeply buried marine sediments on the Peru continental margin using real-time PCR. Environ. Microbiol. 8:1251–1260 - PubMed
-
- Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A, Suzuki M, Takai K, Delwiche M, Colwell FS, Nealson KH, Horikoshi K, D'Hondt S, Jørgensen BB. 2006. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U. S. A. 103:2815–2820 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

