p53's choice of myocardial death or survival: Oxygen protects infarct myocardium by recruiting p53 on NOS3 promoter through regulation of p53-Lys(118) acetylation
- PMID: 24096875
- PMCID: PMC3840484
- DOI: 10.1002/emmm.201202055
p53's choice of myocardial death or survival: Oxygen protects infarct myocardium by recruiting p53 on NOS3 promoter through regulation of p53-Lys(118) acetylation
Abstract
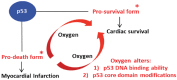
Myocardial infarction, an irreversible cardiac tissue damage, involves progressive loss of cardiomyocytes due to p53-mediated apoptosis. Oxygenation is known to promote cardiac survival through activation of NOS3 gene. We hypothesized a dual role for p53, which, depending on oxygenation, can elicit apoptotic death signals or NOS3-mediated survival signals in the infarct heart. p53 exhibited a differential DNA-binding, namely, BAX-p53RE in the infarct heart or NOS3-p53RE in the oxygenated heart, which was regulated by oxygen-induced, post-translational modification of p53. In the infarct heart, p53 was heavily acetylated at Lys(118) residue, which was exclusively reversed in the oxygenated heart, apparently regulated by oxygen-dependent expression of TIP60. The inhibition of Lys(118) acetylation promoted the generation of NOS3-promoting prosurvival form of p53. Thus, oxygenation switches p53-DNA interaction by regulating p53 core-domain acetylation, promoting a prosurvival transcription activity of p53. Understanding this novel oxygen-p53 survival pathway will open new avenues in cardioprotection molecular therapy.
Keywords: NOS3; lysine acetylation; myocardial infarction; oxygenation; p53.
© 2013 The Authors. Published by John Wiley and Sons, Ltd on behalf of EMBO.
Figures

RT-PCR images show that p53 mRNA is upregulated in both MI and MI + OxCy hearts.
Real-time qPCR values, expressed relative to internal control (IC), show significantly higher levels of p53 mRNA in both MI and MI + OxCy hearts when compared to control. Data represent mean ± SD of five independent measurements. *p = 4.3E−06; #p = 1.5E−09.
Western-blot images show that p53 protein is increased in both MI and MI + OxCy hearts.
In vivo ELISA results show significantly higher levels of p53 in both MI and MI + OxCy hearts when compared to control. Data represent mean ± SD of five independent measurements. *p = 2.0E−06; #p = 5.2E−09.
Immunoprecipitation (IP) data show increase in p53 protein level in MI and MI + OxCy hearts.
Subcellular localization data of p53, obtained by IP of nuclear (NF) and cytoplasmic (CF) fractions from MI and MI + OxCy heart tissues, show that p53 is present in the nuclear fraction of both MI and MI + OxCy hearts.
Binding of p53 to its inhibitor Mdm2 was analysed using co-IP with both anti-p53 and anti-Mdm2 antibodies. The data show that p53 is bound to Mdm2 only in the control hearts, while the MI and MI + OxCy hearts show no interaction of p53 with Mdm2.
p53 transcriptional activity was probed in MI and MI + OxCy hearts by analysing the binding of p53 to its transcriptional activator p300 using co-immunoprecipitation with both anti-p53 and anti-Mdm2 antibodies. The data show that p53 is bound to p300 in both MI and MI + OxCy hearts. Overall, the results established that p53 is upregulated, nuclear, stabilized, and transcriptionally potent in the infarct and oxygenated hearts.

A putative p53 binding site was identified in the NOS3 promoter using Genomatix, MatInspector module. NOS3-p53RE lies between −363 and −341 bp on the 602-bp NOS3 promoter.
pNOS3p-luc1 (NOS3 602-bp promoter luciferase construct) was transfected in L6 cells and the effect of p53 on luciferase activity was measured. Resveratrol was used to activate p53 protein (Western blot). p53 gene-activation induces a 14-fold increase in pNOS3p-luc1 luciferase activity (red bar). p53 gene-silencing using p53 siRNA reverses this effect. Data represent mean ± SD of six independent measurements. *p = 2.0E−12. The results show that p53 transcriptionally regulates NOS3 promoter.
pNOS3p-luc2, the NOS3-p53RE (−363 to −341) cloned in luciferase vector was transfected in L6 cells in presence of resveratrol to activate p53 protein. Results show that p53 induces a 16-fold increase in the NOS3-p53RE luciferase activity (red bar). p53 gene-silencing and p53 transcriptional inhibition using p53 siRNA and pifithrin-α abolishes the increase in luciferase activity. Data represent mean ± SD of six independent measurements. *p = 8.0E−12. The results show that p53 regulates NOS3 promoter via NOS3-p53RE. The mmpNOS3p-luc2 construct (mmp-mutant minimal promoter) with mutated sequence of NOS3-p53RE was transfected in resveratrol-treated L6 cells. No increase in the luciferase activity is observed, showing the specificity of NOS3-p53RE.
Interaction of p53 with NOS3-p53RE is confirmed using EMSA, results show presence of shift and super-shift, p215'RE is used as a positive control.
Chromatin immunoprecipitation (ChIP) was conducted in resveratrol-treated L6 cells to confirm in vivo binding of p53 to NOS3-p53RE. p53 shows an increase in NOS3-p53RE binding with increasing dose of resveratrol. Scrambled primers for PCR were used as negative control.
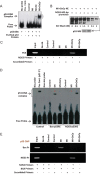
The binding affinity of p53 protein isolated from control (healthy), MI and MI + OxCy hearts towards NOS3-p53RE was analysed using EMSA. p53 proteins isolated from control and MI hearts show no binding to p53RE in NOS3 promoter, while p53 proteins isolated from MI + OxCy hearts show high affinity towards NOS3-p53RE.
Binding affinity of p53 from MI + OxCy was measured using promoter-binding assay. Nuclear extract (NE) of MI + OxCy was incubated with biotin-labelled NOS3-p53RE and the extract was washed using increasing concentration of KCl. Washing of the nuclear extract and biotin-labelled DNA complex with 3M KCl shows a single band in silver staining. The single protein band was eluted from the gel and stained with p53 antibody using Western blotting. The data show that p53 from MI + OxCy hearts have high affinity and specificity for NOS3-p53RE.
ChIP assay was performed to confirm differential binding of p53 to NOS3-p53RE in MI and oxygenated hearts. The data show that p53 does not bind to NOS3-p53RE in healthy and infarcted hearts. On the other hand, oxygenation induces p53 binding to the NOS3-p53RE. Scrambled primers were used as negative control and input was used as a positive loading control.
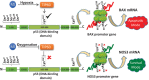
Binding of p53 protein isolated from control, MI and MI + OxCy hearts with NOS3-p53RE and BAX-p53RE was analysed using EMSA. p53 isolated from healthy and MI hearts show no binding to NOS3-p53RE whereas p53 isolated from MI + OxCy hearts show very high binding towards NOS3-p53RE. On other hand, p53 isolated from MI hearts show high binding to p53RE in BAX promoter and p53 binding to the BAX-p53RE drops substantially in MI + OxCy treated hearts.
ChIP assay in these tissues shows similar results where p53 from MI binds to BAX-p53RE and not to NOS3-p53RE. On the other hand, oxygenation induces shift of p53 binding from BAX to NOS3 promoter.

Status of rat p53 post-translational modifications was analysed by immunoprecipitation of p53 with phosphorylation- and acetylation-specific antibodies. Results show that p53 is phosphorylated and acetylated minimally at Ser6, Ser9 and Ser15, and Lys379 in control (healthy) heart. p53 shows very high phosphorylation and acetylation at Ser6, Ser9, Ser15 and Ser20, Thr18 and Lys118, Lys373, and Lys379 residues in the infarct heart. MI + OxCy heart does not show any post-translational modification of p53 from MI heart other than inducing a decrease in acetylation of Lys118 residue.
The role of p53-Lys118 acetylation in the decision of p53 to bind to BAX-p53RE or NOS3-p53RE status was confirmed by incubating the p53 proteins isolated from MI and MI + OxCy hearts with both BAX-p53RE and NOS-p53RE. The bound fraction of p53 was eluted and probed for p53 posttranslational modifications. Results show that p53 from MI heart is mostly bound to BAX-p53RE and this fraction of p53 is acetylated at Lys118 residue. However, a small fraction of p53 from MI heart is also bound to BAX-p53RE but this fraction is not acetylated at Lys118 residue. Similarly p53 from MI + OxCy heart is mostly bound to NOS3-p53RE and this fraction is not acetylated at Lys118. A small fraction of p53 is found bound to BAX-p53RE and this fraction is acetylated at Lys118 residue. The data suggest the importance of Lys118 acetylation in deciding the choice of p53 to bind to either BAX or NOS3 promoter, or in other words, to induce death or survival of cardiomyocytes in the infarct hearts.
The effect of oxygenation upon the transcriptional activity of p53 in upregulating its downstream apoptotic genes were analysed. The activation of 15 apoptotic genes carrying p53 response element was analysed in the healthy, MI and MI + OxCy hearts. The gene-array data show that the p53 downstream genes involved in apoptosis are activated only in the MI hearts. Upon oxygenation, the p53-mediated activation of these genes is significantly inhibited. These genes show minimal expression in the healthy hearts (control).
The expression of 24 anti-apoptotic genes, which might play crucial role in cardiac survival, is observed in the healthy, MI and MI + OxCy hearts. The results show that these genes are minimally expressed in the Infarct hearts (MI). However, upon oxygenation of the MI hearts there is a significant increase in the expression of these genes.
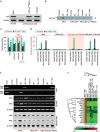
p53 knockout and p53Lys118–Ala118 mutant H9c2 cells were created as described previously (Gogna et al, 2012c). Western-blot analysis shows the knockout of p53 in H9c2 cells (lane 2), while addition of p53Lys118–Ala118 cDNA or p53 Wt cDNA to the H9c2 cells show the expression of p53 (lanes 3,4).
The H9c2, H9c2 p53−/− and H9c2 p53-Lys118(Mut) cells were cultured under serum-deprived (SD) conditions to mimic MI. p53-Lys118 acetylation was analysed in the SD and oxygenated (Oxy) cells by Western blot (WB) and immunoprecipitation (IP) using p53-Lys118 antibody.
The survival potential of H9c2, H9c2 p53−/−, H9c2 p53-Lys118(Mut) and H9c2 p53−/− +p53 Wt-cDNA cells was determined in the control, SD and SD + Oxy treatments. Data represent mean ± SD of eight independent measurements. *p = 6.6E−14 versus respective SD group; #p = 0.0014 versus respective SD group. The results show that oxygenation restored the SD-induced cell death.
Results of luciferase assay showing the activation of p53-NOS3-RE and p53-BAX-RE in H9c2 cells. Oxygenation of SD cells switches from p53-BAX-RE to p53-NOS3-RE activation. Data represent mean ± SD of eight independent measurements.
ChIP assay showing the binding of p53 to its respective NOS3 and BAX RE in H9c2 cells. The data show that p53 binds to BAX-RE in SD cells and shifts its binding to NOS3 upon oxygenation. No binding to either NOS3-RE or BAX-RE is observed in H9c2 p53−/− cells. In H9c2 p53-Lys118(Mut) cells, SD does not induce binding of p53 to the BAX-RE but upon oxygenation p53 binds to the NOS3-RE. The data suggest that p53-Lys118 acetylation is crucial for binding of p53 to BAX-RE and mutation in this site induces p53 binding to the NOS3-RE. Similar results of NOS3 and BAX mRNA and protein upregulation are observed in RT-PCR and the Western-blot assay.
Expression of p53 downstream genes involved in apoptosis in H9c2 cells. The data show that SD results in the activation of these genes, whereas p53−/− and p53-Lys118 (Mut) cells do not activate these genes under any condition.

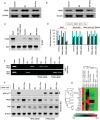
Western-blot results show the abolition of p53 expression in RNC cells (lane 2), while addition of p53Lys118-Ala118 cDNA and p53 Wt cDNA to RNC p53−/− cells show the expression of p53 (lanes 3 and 4). Control cells show expression of p53 (lane 1).
Immunoprecipitation results show acetylation of p53 at Lys118 residue in RNC cells under SD conditions (lane 2). However, this expression is abolished upon oxygenation (lane 3). Acetylation at p53 Lys118 is absent in control, RNC p53−/− and RNC p53-Lys118(Mut) cells under both SD and SD + Oxy conditions (lanes 4–9).
Cell survival analysis of control, SD and SD + Oxy-treated RNC, RNC + p53 siRNA, RNC + p53 siRNA + p53-Lys118(Mut) cDNA and RNC + p53 siRNA + Wt p53 cDNA cells show an increase in the SD-treated RNC + p53 siRNA and RNC + p53 siRNA + p53-Lys118(Mut) cDNA cells compared to RNC cells, exhibiting the crucial role of deacetylation of p53 Lys118 in cell survival. Data represent mean ± SD of eight independent measurements. *p = 2.6E−14 versus respective SD group; #p = 9.3E−08 versus respective SD group.
Luciferase assay shows activation of BAX-p53-RE and NOS3-p53-RE in SD and SD + Oxy-treated RNC cells. Neither BAX nor NOS3 is activated in RNC + p53 siRNA cells. Oxygenation caused a switch of BAX activation to NOS3 activation in RNC p53-Lys118(Mut) cells. Data represent mean ± SD of eight independent measurements.
ChIP assay shows the binding of p53 to its respective NOS3 and BAX RE in RNC cells (lanes 4 and 5). However, no binding of p53 to BAX-RE or NOS3-RE is observed in RNC p53−/− cells (lanes 7 and 8). However, p53 binds to NOS3-RE in SD + Oxy-treated RNC p53-Lys118(Mut) cells (lane 11). These results suggest the importance of p53 Lys118 acetylation in activation of either BAX-RE or NOS3-RE and regulation of apoptotic or survival pathway respectively. Similar results were observed at mRNA and protein level using RT-PCR and Western-blot techniques in RNC, RNC p53−/− and RNC p53-Lys118(Mut) cells.
Expression of p53 downstream genes in RNC, RNC p53−/− and RNC p53-Lys118(Mut) cells. SD-treated RNC cells show activation of apoptotic genes whereas RNC p53−/− and RNC p53-Lys118(Mut) cells show no involvement in apoptotic gene activation.

Expression of p53-interating acetylases in the control, serum-deprived (SD), and oxygenated (SD + Oxy) H9c2, H9c2 p53−/−, H9c2 p53-Lys118(Mut) cells, as determined by in vivo ELISA.
Expression of p53-interating acetylases in the control, serum-deprived (SD), and oxygenated (SD + Oxy) RNC, RNC p53-/-, RNC p53-Lys118(Mut) cells. The results show that the expression of TIP60, a p53-interacting acetylase, is inhibited upon oxygenation in these cells.
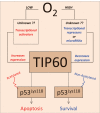
Inhibition of TIP60 expression is also observed in the MI + OxCy heart tissues. Data represent mean ± SD of eight independent measurements in all groups. *p = 6.2E−12 versus respective MI group.
TIP60 protein expression in H9c2 cells under control, SD, and SD + Oxy conditions. SD induces TIP60 expression, which is inhibited by oxygenation.
TIP60 expression in MI hearts is inhibited by oxygenation.
Co-immunoprecipitation of p53 with TIP60 shows that TIP60 binds to p53 only in MI hearts and this interaction is abolished upon oxygenation. Input and SD-treated H9c2 cells with TIP60 gene-silencing were used as controls.
Effect of TIP60 on the survival of SD H9c2 cells. Silencing of TIP60 using TIP60 siRNA in H9c2, H9c2 p53−/− and H9c2 p53-Lys118(Mut) cells increases the survival of SD cells. Addition of TIP60 using cDNA results decreases the survival of SD cells. Data represent mean ± SD of four independent measurements.
Effect of TIP60 on the binding of p53 to BAX-RE and NOS3-RE was analysed using ChIP in H9c2 cells. Results show that TIP60 gene-silencing results in binding of p53 to NOS3-RE in both SD and SD + Oxy cells. Similarly, exogenous addition of TIP60 cDNA results in binding of p53 to BAX-RE in both SD and SD + Oxy cells.
Effect of TIP60 siRNA and TIP60 cDNA addition on p53-Lys118 acetylation, NOS3 and BAX protein expression was determined using Western blot. Data show that TIP60 siRNA abolishes p53-Lys118 acetylation, increases the expression of NOS3 protein and decreases BAX expression in H9c2 cells. Similarly, TIP60 cDNA increases p53-Lys118 acetylation, decreases the expression of NOS3 protein and increased BAX expression in H9c2 cells.
Expression of p53 downstream genes was determined in the SD and SD + Oxy group of H9c2 cells in presence and absence of TIP60 siRNA and TIP60 cDNA. Data show that TIP60 cDNA increases the expression of these genes and TIP60 siRNA decreases their expression in both SD and SD + Oxy groups.
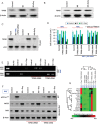
Western-blot analysis was conducted to study the expression of TIP60 in control, SD and SD + Oxy-treated cells. SD induces TIP60 expression (lane 2); however, this expression is significantly inhibited by oxygenation (lane 3).
Western blots of TIP60 expression in MI and MI + Oxy-treated cardiac tissue. Results show increased TIP60 expression in MI tissue (lane 2); however, this expression is significantly reduced in MI + Oxy-treated cardiac tissue (lane 3).
Co-immunoprecipitation study was conducted to study p53 and TIP60 interaction in MI and MI + Oxy-treated RNC cells. Results show p53 interaction with TIP60 in SD-treated cells (lane 4); however, this oxygenation significantly inhibits this interaction (lane 5) in RNC cells. Input and TIP60 siRNA were used as controls for the study.
The role of TIP60 in cell survival was studied in RNC cells using flow cytometry. Transfection with TIP60 siRNA results in increase in cell-survival fraction in RNC cells while TIP60 cDNA results in a decrease in cell-survival fraction. Conversely, neither TIP60 siRNA nor TIP60 cDNA has any effect on cell survival in RNC p53−/− and RNC p53-Lys118(Mut) cells. Data represent mean ± SD of four independent measurements.
CHIP assay was conducted to study the role of TIP60 in p53 binding to BAX-RE or NOS3-RE in SD and SD + Oxy-treated RNC cells. Results show that transfection with TIP60 siRNA results in p53 binding to NOS-RE in both SD and SD + Oxy-treated cells. However, exogenous addition of TIP60 cDNA result in p53 binding to BAX-RE in both SDF and SD + Oxy-treated cells.
Effect of TIP60 siRNA and TIP60 cDNA addition on p53-Lys118 acetylation, NOS3 and BAX protein expression was studied in SD and SD + Oxy-treated RNC cells. Transfection with TIP60 siRNA led to abolition of p53-Lys118 acetylation, increased NOS3 expression and decreased BAX expression in RNC cells. Contrarily, transfection with TIP60 cDNA led to increased p53-Lys118 acetylation, decreased NOS3 expression and increased BAX expression in RNC cells.
Gene array was studied to study the role of TIP60 in activation of p53 downstream genes in SD and SD + Oxy treated cells. Transfection with TIP60 cDNA results in the activations of apoptotic genes in SD and SD + Oxy-treated cells when compared to TIP60 siRNA-transfected SD and SD + Oxy-treated groups.
References
-
- Alvarado-Vasquez N, Zapata E, Alcazar-Leyva S, Masso F, Montano LF. Reduced NO synthesis and eNOS mRNA expression in endothelial cells from newborns with a strong family history of type 2 diabetes. Diabetes Metab Res Rev. 2007;23:559–566. - PubMed
-
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–1254. - PubMed
-
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

