Conserved pyridoxal protein that regulates Ile and Val metabolism
- PMID: 24097949
- PMCID: PMC3889608
- DOI: 10.1128/JB.00593-13
Conserved pyridoxal protein that regulates Ile and Val metabolism
Abstract
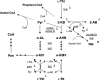
Escherichia coli YggS is a member of the highly conserved uncharacterized protein family that binds pyridoxal 5'-phosphate (PLP). To assist with the functional assignment of the YggS family, in vivo and in vitro analyses were performed using a yggS-deficient E. coli strain (ΔyggS) and a purified form of YggS, respectively. In the stationary phase, the ΔyggS strain exhibited a completely different intracellular pool of amino acids and produced a significant amount of l-Val in the culture medium. The log-phase ΔyggS strain accumulated 2-ketobutyrate, its aminated compound 2-aminobutyrate, and, to a lesser extent, l-Val. It also exhibited a 1.3- to 2.6-fold increase in the levels of Ile and Val metabolic enzymes. The fact that similar phenotypes were induced in wild-type E. coli by the exogenous addition of 2-ketobutyrate and 2-aminobutyrate indicates that the 2 compounds contribute to the ΔyggS phenotypes. We showed that the initial cause of the keto acid imbalance was the reduced availability of coenzyme A (CoA); supplementation with pantothenate, which is a CoA precursor, fully reversed phenotypes conferred by the yggS mutation. The plasmid-borne expression of YggS and orthologs from Bacillus subtilis, Saccharomyces cerevisiae, and humans fully rescued the ΔyggS phenotypes. Expression of a mutant YggS lacking PLP-binding ability, however, did not reverse the ΔyggS phenotypes. These results demonstrate for the first time that YggS controls Ile and Val metabolism by modulating 2-ketobutyrate and CoA availability. Its function depends on PLP, and it is highly conserved in a wide range species, from bacteria to humans.
Figures
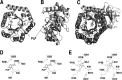
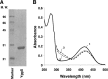



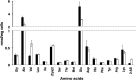
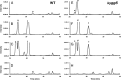


References
-
- Ito T, Uozumi N, Nakamura T, Takayama S, Matsuda N, Aiba H, Hemmi H, Yoshimura T. 2009. The implication of YggT of Escherichia coli in osmotic regulation. Biosci. Biotechnol. Biochem. 73:2698–2704 - PubMed
-
- Bradshaw JS, Kuzminov A. 2003. RdgB acts to avoid chromosome fragmentation in Escherichia coli. Mol. Microbiol. 48:1711–1725 - PubMed
-
- Eswaramoorthy S, Gerchman S, Graziano V, Kycia H, Studier FW, Swaminathan S. 2003. Structure of a yeast hypothetical protein selected by a structural genomics approach. Acta Crystallogr. D Biol. Crystallogr. 59:127–135 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

