Identification and characterization of novel Helicobacter pylori apo-fur-regulated target genes
- PMID: 24097951
- PMCID: PMC3889615
- DOI: 10.1128/JB.01026-13
Identification and characterization of novel Helicobacter pylori apo-fur-regulated target genes
Abstract
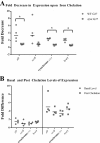
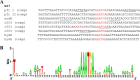
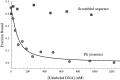
In Helicobacter pylori, the ferric uptake regulator (Fur) has evolved additional regulatory functions not seen in other bacteria; it can repress and activate different groups of genes in both its iron-bound and apo forms. Because little is understood about the process of apo-Fur repression and because only two apo-Fur-repressed genes (pfr and sodB) have previously been identified, we sought to expand our understanding of this type of regulation. Utilizing published genomic studies, we selected three potential new apo-Fur-regulated gene targets: serB, hydA, and the cytochrome c553 gene. Transcriptional analyses confirmed Fur-dependent repression of these genes in the absence of iron, as well as derepression in the absence of Fur. Binding studies showed that apo-Fur directly interacted with the suspected hydA and cytochrome c553 promoters but not that of serB, which was subsequently shown to be cotranscribed with pfr; apo-Fur-dependent regulation occurred at the pfr promoter. Alignments of apo-regulated promoter regions revealed a conserved, 6-bp consensus sequence (AAATGA). DNase I footprinting showed that this sequence lies within the protected regions of the pfr and hydA promoters. Moreover, mutation of the sequence in the pfr promoter abrogated Fur binding and DNase protection. Likewise, fluorescence anisotropy studies and binding studies with mutated consensus sequences showed that the sequence was important for apo-Fur binding to the pfr promoter. Together these studies expand the known apo-Fur regulon in H. pylori and characterize the first reported apo-Fur box sequence.
Figures
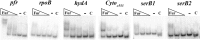





References
-
- Blaser MJ. 1990. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J. Infect. Dis. 161:626–633 - PubMed
-
- Asaka M, Kudo M, Kato M, Sugiyama T, Takeda H. 1998. Review article: long-term Helicobacter pylori infection—from gastritis to gastric cancer. Aliment. Pharmacol. Ther. 12(Suppl 1):9–15 - PubMed
-
- Delany I, Spohn G, Rappuoli R, Scarlato V. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297–1309 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

