Ligand clouds around protein clouds: a scenario of ligand binding with intrinsically disordered proteins
- PMID: 24098099
- PMCID: PMC3789766
- DOI: 10.1371/journal.pcbi.1003249
Ligand clouds around protein clouds: a scenario of ligand binding with intrinsically disordered proteins
Abstract
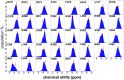
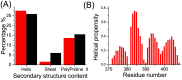
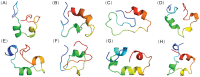
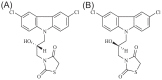
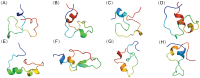
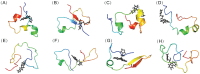
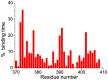
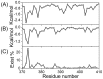
Intrinsically disordered proteins (IDPs) were found to be widely associated with human diseases and may serve as potential drug design targets. However, drug design targeting IDPs is still in the very early stages. Progress in drug design is usually achieved using experimental screening; however, the structural disorder of IDPs makes it difficult to characterize their interaction with ligands using experiments alone. To better understand the structure of IDPs and their interactions with small molecule ligands, we performed extensive simulations on the c-Myc₃₇₀₋₄₀₉ peptide and its binding to a reported small molecule inhibitor, ligand 10074-A4. We found that the conformational space of the apo c-Myc₃₇₀₋₄₀₉ peptide was rather dispersed and that the conformations of the peptide were stabilized mainly by charge interactions and hydrogen bonds. Under the binding of the ligand, c-Myc₃₇₀₋₄₀₉ remained disordered. The ligand was found to bind to c-Myc₃₇₀₋₄₀₉ at different sites along the chain and behaved like a 'ligand cloud'. In contrast to ligand binding to more rigid target proteins that usually results in a dominant bound structure, ligand binding to IDPs may better be described as ligand clouds around protein clouds. Nevertheless, the binding of the ligand and a non-ligand to the c-Myc₃₇₀₋₄₀₉ target could be clearly distinguished. The present study provides insights that will help improve rational drug design that targets IDPs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


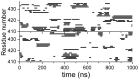
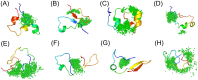
References
-
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z (2002) Intrinsic disorder and protein function. Biochemistry 41: 6573–6582. - PubMed
-
- Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208. - PubMed
-
- Tompa P (2002) Intrinsically unstructured proteins. Trends Biochem Sci 27: 527–533. - PubMed
-
- Huang Y, Liu Z (2010) Intrinsically disordered proteins: the new sequence-structure-function relations. Acta Phys Chim Sin 26: 2061–2072.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

