Herpes simplex virus type 1 single strand DNA binding protein and helicase/primase complex disable cellular ATR signaling
- PMID: 24098119
- PMCID: PMC3789782
- DOI: 10.1371/journal.ppat.1003652
Herpes simplex virus type 1 single strand DNA binding protein and helicase/primase complex disable cellular ATR signaling
Abstract
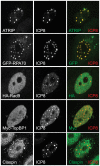
Herpes Simplex Virus type 1 (HSV-1) has evolved to disable the cellular DNA damage response kinase, ATR. We have previously shown that HSV-1-infected cells are unable to phosphorylate the ATR substrate Chk1, even under conditions in which replication forks are stalled. Here we report that the HSV-1 single stranded DNA binding protein (ICP8), and the helicase/primase complex (UL8/UL5/UL52) form a nuclear complex in transfected cells that is necessary and sufficient to disable ATR signaling. This complex localizes to sites of DNA damage and colocalizes with ATR/ATRIP and RPA, but under these conditions, the Rad9-Rad1-Hus1 checkpoint clamp (9-1-1) do not. ATR is generally activated by substrates that contain ssDNA adjacent to dsDNA, and previous work from our laboratory has shown that ICP8 and helicase/primase also recognize this substrate. We suggest that these four viral proteins prevent ATR activation by binding to the DNA substrate and obstructing loading of the 9-1-1 checkpoint clamp. Exclusion of 9-1-1 prevents recruitment of TopBP1, the ATR kinase activator, and thus effectively disables ATR signaling. These data provide the first example of viral DNA replication proteins obscuring access to a DNA substrate that would normally trigger a DNA damage response and checkpoint signaling. This unusual mechanism used by HSV suggests that it may be possible to inhibit ATR signaling by preventing recruitment of the 9-1-1 clamp and TopBP1.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

