Alternative splicing targeting the hTAF4-TAFH domain of TAF4 represses proliferation and accelerates chondrogenic differentiation of human mesenchymal stem cells
- PMID: 24098348
- PMCID: PMC3788782
- DOI: 10.1371/journal.pone.0074799
Alternative splicing targeting the hTAF4-TAFH domain of TAF4 represses proliferation and accelerates chondrogenic differentiation of human mesenchymal stem cells
Abstract
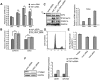
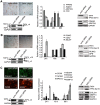
Transcription factor IID (TFIID) activity can be regulated by cellular signals to specifically alter transcription of particular subsets of genes. Alternative splicing of TFIID subunits is often the result of external stimulation of upstream signaling pathways. We studied tissue distribution and cellular expression of different splice variants of TFIID subunit TAF4 mRNA and biochemical properties of its isoforms in human mesenchymal stem cells (hMSCs) to reveal the role of different isoforms of TAF4 in the regulation of proliferation and differentiation. Expression of TAF4 transcripts with exons VI or VII deleted, which results in a structurally modified hTAF4-TAFH domain, increases during early differentiation of hMSCs into osteoblasts, adipocytes and chondrocytes. Functional analysis data reveals that TAF4 isoforms with the deleted hTAF4-TAFH domain repress proliferation of hMSCs and preferentially promote chondrogenic differentiation at the expense of other developmental pathways. This study also provides initial data showing possible cross-talks between TAF4 and TP53 activity and switching between canonical and non-canonical WNT signaling in the processes of proliferation and differentiation of hMSCs. We propose that TAF4 isoforms generated by the alternative splicing participate in the conversion of the cellular transcriptional programs from the maintenance of stem cell state to differentiation, particularly differentiation along the chondrogenic pathway.
Conflict of interest statement
Figures


References
-
- Müller F, Zaucker A, Tora L (2010) Developmental regulation of transcription initiation: more than just changing the actors. Curr Opin Genet Dev 20: 533–40. - PubMed
-
- Burley SK, Roeder RG (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem 65: 769–99. - PubMed
-
- Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, et al.. (2013) The architecture of human general transcription factor TFIID core complex. Nature. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

