Transcriptome comparison of human neurons generated using induced pluripotent stem cells derived from dental pulp and skin fibroblasts
- PMID: 24098394
- PMCID: PMC3789755
- DOI: 10.1371/journal.pone.0075682
Transcriptome comparison of human neurons generated using induced pluripotent stem cells derived from dental pulp and skin fibroblasts
Abstract
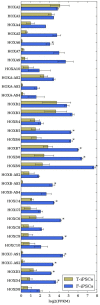
Induced pluripotent stem cell (iPSC) technology is providing an opportunity to study neuropsychiatric disorders through the capacity to grow patient-specific neurons in vitro. Skin fibroblasts obtained by biopsy have been the most reliable source of cells for reprogramming. However, using other somatic cells obtained by less invasive means would be ideal, especially in children with autism spectrum disorders (ASD) and other neurodevelopmental conditions. In addition to fibroblasts, iPSCs have been developed from cord blood, lymphocytes, hair keratinocytes, and dental pulp from deciduous teeth. Of these, dental pulp would be a good source for neurodevelopmental disorders in children because obtaining material is non-invasive. We investigated its suitability for disease modeling by carrying out gene expression profiling, using RNA-seq, on differentiated neurons derived from iPSCs made from dental pulp extracted from deciduous teeth (T-iPSCs) and fibroblasts (F-iPSCs). This is the first RNA-seq analysis comparing gene expression profiles in neurons derived from iPSCs made from different somatic cells. For the most part, gene expression profiles were quite similar with only 329 genes showing differential expression at a nominally significant p-value (p<0.05), of which 63 remained significant after correcting for genome-wide analysis (FDR <0.05). The most striking difference was the lower level of expression detected for numerous members of the all four HOX gene families in neurons derived from T-iPSCs. In addition, an increased level of expression was seen for several transcription factors expressed in the developing forebrain (FOXP2, OTX1, and LHX2, for example). Overall, pathway analysis revealed that differentially expressed genes that showed higher levels of expression in neurons derived from T-iPSCs were enriched for genes implicated in schizophrenia (SZ). The findings suggest that neurons derived from T-iPSCs are suitable for disease-modeling neuropsychiatric disorder and may have some advantages over those derived from F-iPSCs.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- MH073164/MH/NIMH NIH HHS/United States
- KL2 RR025749/RR/NCRR NIH HHS/United States
- R21 MH087840/MH/NIMH NIH HHS/United States
- R01 MH073164/MH/NIMH NIH HHS/United States
- TL1 RR025748/RR/NCRR NIH HHS/United States
- MH087840/MH/NIMH NIH HHS/United States
- R33 MH087840/MH/NIMH NIH HHS/United States
- 1P30HD071593-01/HD/NICHD NIH HHS/United States
- KL2RR025749/RR/NCRR NIH HHS/United States
- UL1 RR025750/RR/NCRR NIH HHS/United States
- UL1RR025750/RR/NCRR NIH HHS/United States
- P30 HD071593/HD/NICHD NIH HHS/United States
- TL1RR025748/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

