DNA methylation mediates persistent epileptiform activity in vitro and in vivo
- PMID: 24098468
- PMCID: PMC3788713
- DOI: 10.1371/journal.pone.0076299
DNA methylation mediates persistent epileptiform activity in vitro and in vivo
Abstract
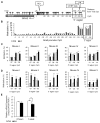
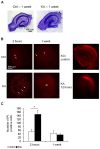
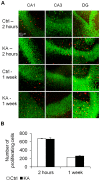
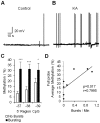
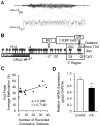
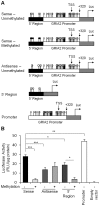
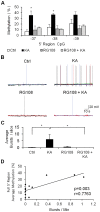
Epilepsy is a chronic brain disorder involving recurring seizures often precipitated by an earlier neuronal insult. The mechanisms that link the transient neuronal insult to the lasting state of epilepsy are unknown. Here we tested the possible role of DNA methylation in mediating long-term induction of epileptiform activity by transient kainic acid exposure using in vitro and in vivo rodent models. We analyzed changes in the gria2 gene, which encodes for the GluA2 subunit of the ionotropic glutamate, alpha-amino-3-hydroxy-5-methyl-4-isoxazole proprionic acid receptor and is well documented to play a role in epilepsy. We show that kainic acid exposure for two hours to mouse hippocampal slices triggers methylation of a 5' regulatory region of the gria2 gene. Increase in methylation persists one week after removal of the drug, with concurrent suppression of gria2 mRNA expression levels. The degree of kainic acid-induced hypermethylation of gria2 5' region varies between individual slices and correlates with the changes in excitability induced by kainic acid. In a rat in vivo model of post kainic acid-induced epilepsy, we show similar hypermethylation of the 5' region of gria2. Inter-individual variations in gria2 methylation, correlate with the frequency and intensity of seizures among epileptic rats. Luciferase reporter assays support a regulatory role for methylation of gria2 5' region. Inhibition of DNA methylation by RG108 blocked kainic acid-induced hypermethylation of gria2 5' region in hippocampal slice cultures and bursting activity. Our results suggest that DNA methylation of such genes as gria2 mediates persistent epileptiform activity and inter-individual differences in the epileptic response to neuronal insult and that pharmacological agents that block DNA methylation inhibit epileptiform activity raising the prospect of DNA methylation inhibitors in epilepsy therapeutics.
Conflict of interest statement
Figures
References
-
- Razin A (1998) CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J 17: 4905-4908. doi:10.1093/emboj/17.17.4905. PubMed: 9724627. - DOI - PMC - PubMed
-
- Szyf M (2009) Epigenetics, DNA Methylation, and Chromatin Modifying Drugs. Annu Rev Pharmacol Toxicol 49: 243-263. doi:10.1146/annurev-pharmtox-061008-103102. PubMed: 18851683. - DOI - PubMed
-
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S et al. (2004) Epigenetic programming by maternal behavior. Nat Neurosci 7: 847-854. doi:10.1038/nn1276. PubMed: 15220929. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

