Arginine methylation of hnRNP A2 does not directly govern its subcellular localization
- PMID: 24098712
- PMCID: PMC3787039
- DOI: 10.1371/journal.pone.0075669
Arginine methylation of hnRNP A2 does not directly govern its subcellular localization
Abstract
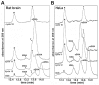
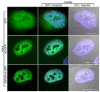
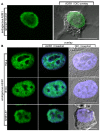
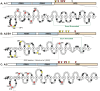
The hnRNP A/B paralogs A1, A2/B1 and A3 are key components of the nuclear 40S hnRNP core particles. Despite a high degree of sequence similarity, increasing evidence suggests they perform additional, functionally distinct roles in RNA metabolism. Here we identify and study the functional consequences of differential post-translational modification of hnRNPs A1, A2 and A3. We show that while arginine residues in the RGG box domain of hnRNP A1 and A3 are almost exhaustively, asymmetrically dimethylated, hnRNP A2 is dimethylated at only a single residue (Arg-254) and this modification is conserved across cell types. It has been suggested that arginine methylation regulates the nucleocytoplasmic distribution of hnRNP A/B proteins. However, we show that transfected cells expressing an A2(R254A) point mutant exhibit no difference in subcellular localization. Similarly, immunostaining and mass spectrometry of endogenous hnRNP A2 in transformed cells reveals a naturally-occurring pool of unmethylated protein but an exclusively nuclear pattern of localization. Our results suggest an alternative role for post-translational arginine methylation of hnRNPs and offer further evidence that the hnRNP A/B paralogs are not functionally redundant.
Conflict of interest statement
Figures



Similar articles
-
The RGG domain in hnRNP A2 affects subcellular localization.Exp Cell Res. 2000 May 1;256(2):522-32. doi: 10.1006/excr.2000.4827. Exp Cell Res. 2000. PMID: 10772824
-
Methylarginines within the RGG-Motif Region of hnRNP A1 Affect Its IRES Trans-Acting Factor Activity and Are Required for hnRNP A1 Stress Granule Localization and Formation.J Mol Biol. 2017 Jan 20;429(2):295-307. doi: 10.1016/j.jmb.2016.12.011. Epub 2016 Dec 13. J Mol Biol. 2017. PMID: 27979648
-
Proteomic analysis of methylarginine-containing proteins in HeLa cells by two-dimensional gel electrophoresis and immunoblotting with a methylarginine-specific antibody.Protein J. 2009 May;28(3-4):139-47. doi: 10.1007/s10930-009-9174-3. Protein J. 2009. PMID: 19365714
-
Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: from structural insights to post-transcriptional regulatory roles.Mol Cell Neurosci. 2013 Sep;56:436-46. doi: 10.1016/j.mcn.2012.12.002. Epub 2012 Dec 14. Mol Cell Neurosci. 2013. PMID: 23247072 Review.
-
Recent advances in protein methylation: enzymatic methylation of nucleic acid binding proteins.Amino Acids. 1998;15(4):291-306. doi: 10.1007/BF01320895. Amino Acids. 1998. PMID: 9891755 Review.
Cited by
-
hnRNP A/B Proteins: An Encyclopedic Assessment of Their Roles in Homeostasis and Disease.Biology (Basel). 2021 Jul 24;10(8):712. doi: 10.3390/biology10080712. Biology (Basel). 2021. PMID: 34439945 Free PMC article. Review.
-
Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation.Mol Cell. 2018 Feb 1;69(3):465-479.e7. doi: 10.1016/j.molcel.2017.12.022. Epub 2018 Jan 18. Mol Cell. 2018. PMID: 29358076 Free PMC article.
-
Cellular consequences of arginine methylation.Cell Mol Life Sci. 2019 Aug;76(15):2933-2956. doi: 10.1007/s00018-019-03140-2. Epub 2019 May 17. Cell Mol Life Sci. 2019. PMID: 31101937 Free PMC article. Review.
-
PRMT5 regulates IRES-dependent translation via methylation of hnRNP A1.Nucleic Acids Res. 2017 May 5;45(8):4359-4369. doi: 10.1093/nar/gkw1367. Nucleic Acids Res. 2017. PMID: 28115626 Free PMC article.
-
The Role of RNA Splicing Factors in Cancer: Regulation of Viral and Human Gene Expression in Human Papillomavirus-Related Cervical Cancer.Front Cell Dev Biol. 2020 Jun 12;8:474. doi: 10.3389/fcell.2020.00474. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32596243 Free PMC article. Review.
References
-
- Beyer AL, Miller OL, McKnight SL (1980) Ribonucleoprotein structure in nascent hnRNA is nonrandom and sequence-dependent. Cell 20: 75–84. - PubMed
-
- Beyer AL, Christensen ME, Walker BW, Le Stourgeon WM (1977) Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell 11: 127–138. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

