2mit, an intronic gene of Drosophila melanogaster timeless2, is involved in behavioral plasticity
- PMID: 24098788
- PMCID: PMC3786989
- DOI: 10.1371/journal.pone.0076351
2mit, an intronic gene of Drosophila melanogaster timeless2, is involved in behavioral plasticity
Abstract
Background: Intronic genes represent ~6% of the total gene complement in Drosophila melanogaster and ~85% of them encode for proteins. We recently characterized the D. melanogaster timeless2 (tim2) gene, showing its active involvement in chromosomal stability and light synchronization of the adult circadian clock. The protein coding gene named 2mit maps on the 11(th) tim2 intron in the opposite transcriptional orientation.
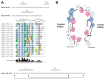
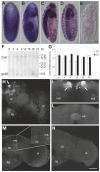
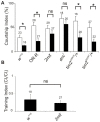
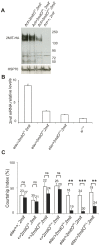
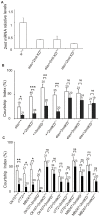
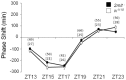
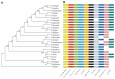
Methodology/principal findings: Here we report the molecular and functional characterization of 2mit. The 2mit gene is expressed throughout Drosophila development, localizing mainly in the nervous system during embryogenesis and mostly in the mushroom bodies and ellipsoid body of the central complex in the adult brain. In silico analyses revealed that 2mit encodes a putative leucine-Rich Repeat transmembrane receptor with intrinsically disordered regions, harboring several fully conserved functional interaction motifs in the cytosolic side. Using insertional mutations, tissue-specific over-expression, and down-regulation approaches, it was found that 2mit is implicated in adult short-term memory, assessed by a courtship conditioning assay. In D. melanogaster, tim2 and 2mit do not seem to be functionally related. Bioinformatic analyses identified 2MIT orthologs in 21 Drosophilidae, 4 Lepidoptera and in Apis mellifera. In addition, the tim2-2mit host-nested gene organization was shown to be present in A. mellifera and maintained among Drosophila species. Within the Drosophilidae 2mit-hosting tim2 intron, in silico approaches detected a neuronal specific transcriptional binding site which might have contributed to preserve the specific host-nested gene association across Drosophila species.
Conclusions/significance: Taken together, these results indicate that 2mit, a gene mainly expressed in the nervous system, has a role in the behavioral plasticity of the adult Drosophila. The presence of a putative 2mit regulatory enhancer within the 2mit-hosting tim2 intron could be considered an evolutionary constraint potentially involved in maintaining the tim2-2mit host-nested chromosomal architecture during the evolution of Drosophila species.
Conflict of interest statement
Figures

References
-
- Bhutkar A, Russo SM, Smith TF, Gelbart WM (2007) Genome-scale analysis of positionally relocated genes. Genome Res 17: 1880-1887. doi:10.1101/gr.7062307. PubMed: 17989252. - DOI - PMC - PubMed
-
- Kumar A (2009) An overview of nested genes in eukaryotic genomes. Eukaryot Cell 8: 1321-1329. doi:10.1128/EC.00143-09. PubMed: 19542305. - DOI - PMC - PubMed
-
- Assis R, Kondrashov AS, Koonin EV, Kondrashov FA (2008) Nested genes and increasing organizational complexity of metazoan genomes. Trends Genet 24: 475-478. doi:10.1016/j.tig.2008.08.003. PubMed: 18774620. - DOI - PMC - PubMed
-
- Gibson CW, Thomson NH, Abrams WR, Kirkham J (2005) Nested genes: biological implications and use of AFM for analysis. Gene 350: 15-23. doi:10.1016/j.gene.2004.12.045. PubMed: 15780979. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

