Common mechanistic themes for the powerstroke of kinesin-14 motors
- PMID: 24099757
- PMCID: PMC3851574
- DOI: 10.1016/j.jsb.2013.09.020
Common mechanistic themes for the powerstroke of kinesin-14 motors
Abstract
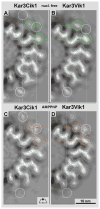
Kar3Cik1 is a heterodimeric kinesin-14 from Saccharomyces cerevisiae involved in spindle formation during mitosis and karyogamy in mating cells. Kar3 represents a canonical kinesin motor domain that interacts with microtubules under the control of ATP-hydrolysis. In vivo, the localization and function of Kar3 is differentially regulated by its interacting stoichiometrically with either Cik1 or Vik1, two closely related motor homology domains that lack the nucleotide-binding site. Indeed, Vik1 structurally resembles the core of a kinesin head. Despite being closely related, Kar3Cik1 and Kar3Vik1 are each responsible for a distinct set of functions in vivo and also display different biochemical behavior in vitro. To determine a structural basis for their distinct functional abilities, we used cryo-electron microscopy and helical reconstruction to investigate the 3-D structure of Kar3Cik1 complexed to microtubules in various nucleotide states and compared our 3-D data of Kar3Cik1 with that of Kar3Vik1 and the homodimeric kinesin-14 Ncd from Drosophila melanogaster. Due to the lack of an X-ray crystal structure of the Cik1 motor homology domain, we predicted the structure of this Cik1 domain based on sequence similarity to its relatives Vik1, Kar3 and Ncd. By molecular docking into our 3-D maps, we produced a detailed near-atomic model of Kar3Cik1 complexed to microtubules in two distinct nucleotide states, a nucleotide-free state and an ATP-bound state. Our data show that despite their functional differences, heterodimeric Kar3Cik1 and Kar3Vik1 and homodimeric Ncd, all share striking structural similarities at distinct nucleotide states indicating a common mechanistic theme within the kinesin-14 family.
Keywords: Cryo-electron microscopy; Helical 3-D analysis; Kar3Cik1; Kinesin-14; Microtubules; Molecular docking.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures






Similar articles
-
Drosophila Ncd reveals an evolutionarily conserved powerstroke mechanism for homodimeric and heterodimeric kinesin-14s.Proc Natl Acad Sci U S A. 2015 May 19;112(20):6359-64. doi: 10.1073/pnas.1505531112. Epub 2015 May 4. Proc Natl Acad Sci U S A. 2015. PMID: 25941402 Free PMC article.
-
The ATPase pathway that drives the kinesin-14 Kar3Vik1 powerstroke.J Biol Chem. 2012 Oct 26;287(44):36673-82. doi: 10.1074/jbc.M112.395590. Epub 2012 Sep 12. J Biol Chem. 2012. PMID: 22977241 Free PMC article.
-
Kar3Vik1, a member of the kinesin-14 superfamily, shows a novel kinesin microtubule binding pattern.J Cell Biol. 2012 Jun 25;197(7):957-70. doi: 10.1083/jcb.201201132. J Cell Biol. 2012. PMID: 22734002 Free PMC article.
-
Interaction of kinesin motors, microtubules, and MAPs.J Muscle Res Cell Motil. 2006;27(2):125-37. doi: 10.1007/s10974-005-9051-4. Epub 2005 Dec 17. J Muscle Res Cell Motil. 2006. PMID: 16362723 Review.
-
Structures of kinesin and kinesin-microtubule interactions.Curr Opin Cell Biol. 1999 Feb;11(1):34-44. doi: 10.1016/s0955-0674(99)80005-2. Curr Opin Cell Biol. 1999. PMID: 10047529 Review.
Cited by
-
Drosophila Ncd reveals an evolutionarily conserved powerstroke mechanism for homodimeric and heterodimeric kinesin-14s.Proc Natl Acad Sci U S A. 2015 May 19;112(20):6359-64. doi: 10.1073/pnas.1505531112. Epub 2015 May 4. Proc Natl Acad Sci U S A. 2015. PMID: 25941402 Free PMC article.
-
Non-catalytic motor domains enable processive movement and functional diversification of the kinesin-14 Kar3.Elife. 2015 Jan 27;4:e04489. doi: 10.7554/eLife.04489. Elife. 2015. PMID: 25626168 Free PMC article.
-
Structural switching of tubulin in the microtubule lattice.Biochem Soc Trans. 2025 Feb 5;53(1):BST20240360. doi: 10.1042/BST20240360. Biochem Soc Trans. 2025. PMID: 39910801 Free PMC article. Review.
-
Kar3Vik1 mechanochemistry is inhibited by mutation or deletion of the C terminus of the Vik1 subunit.J Biol Chem. 2013 Dec 27;288(52):36957-70. doi: 10.1074/jbc.M113.492264. Epub 2013 Nov 16. J Biol Chem. 2013. PMID: 24240171 Free PMC article.
References
-
- Arnal I, Metoz F, DeBonis S, Wade RH. Three-dimensional structure of functional motor proteins on microtubules. Curr Biol. 1996;6:1265–70. - PubMed
-
- Beuron F, Hoenger A. Structural analysis of the microtubule-kinesin complex by cryo-electron microscopy. Methods Mol Biol. 2001;164:235–54. - PubMed
-
- Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 1997;90:959–966. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

