Cell-specific actions of a human LHX3 gene enhancer during pituitary and spinal cord development
- PMID: 24100213
- PMCID: PMC3857196
- DOI: 10.1210/me.2013-1161
Cell-specific actions of a human LHX3 gene enhancer during pituitary and spinal cord development
Abstract
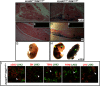
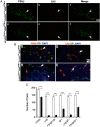
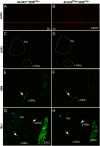
The LIM class of homeodomain protein 3 (LHX3) transcription factor is essential for pituitary gland and nervous system development in mammals. In humans, mutations in the LHX3 gene underlie complex pediatric syndromes featuring deficits in anterior pituitary hormones and defects in the nervous system. The mechanisms that control temporal and spatial expression of the LHX3 gene are poorly understood. The proximal promoters of the human LHX3 gene are insufficient to guide expression in vivo and downstream elements including a conserved enhancer region appear to play a role in tissue-specific expression in the pituitary and nervous system. Here we characterized the activity of this downstream enhancer region in regulating gene expression at the cellular level during development. Human LHX3 enhancer-driven Cre reporter transgenic mice were generated to facilitate studies of enhancer actions. The downstream LHX3 enhancer primarily guides gene transcription in α-glycoprotein subunit -expressing cells secreting the TSHβ, LHβ, or FSHβ hormones and expressing the GATA2 and steroidogenic factor 1 transcription factors. In the developing nervous system, the enhancer serves as a targeting module active in V2a interneurons. These results demonstrate that the downstream LHX3 enhancer is important in specific endocrine and neural cell types but also indicate that additional regulatory elements are likely involved in LHX3 gene expression. Furthermore, these studies revealed significant gonadotrope cell heterogeneity during pituitary development, providing insights into the cellular physiology of this key reproductive regulatory cell. The human LHX3 enhancer-driven Cre reporter transgenic mice also provide a valuable tool for further developmental studies of cell determination and differentiation in the pituitary and nervous system.
Figures





Similar articles
-
A distal modular enhancer complex acts to control pituitary- and nervous system-specific expression of the LHX3 regulatory gene.Mol Endocrinol. 2012 Feb;26(2):308-19. doi: 10.1210/me.2011-1252. Epub 2011 Dec 22. Mol Endocrinol. 2012. PMID: 22194342 Free PMC article.
-
Lhx4 surpasses its paralog Lhx3 in promoting the differentiation of spinal V2a interneurons.Cell Mol Life Sci. 2024 Jul 6;81(1):286. doi: 10.1007/s00018-024-05316-x. Cell Mol Life Sci. 2024. PMID: 38970652 Free PMC article.
-
GATA2-induced silencing and LIM-homeodomain protein-induced activation are mediated by a bi-functional response element in the rat GnRH receptor gene.Mol Endocrinol. 2013 Jan;27(1):74-91. doi: 10.1210/me.2012-1182. Epub 2012 Dec 4. Mol Endocrinol. 2013. PMID: 23211524 Free PMC article.
-
Roles of the LHX3 and LHX4 LIM-homeodomain factors in pituitary development.Mol Cell Endocrinol. 2007 Feb;265-266:190-5. doi: 10.1016/j.mce.2006.12.019. Epub 2007 Jan 8. Mol Cell Endocrinol. 2007. PMID: 17210222 Free PMC article. Review.
-
Possible role of PACAP and its PAC1 receptor in the differential regulation of pituitary LHbeta- and FSHbeta-subunit gene expression by pulsatile GnRH stimulation.Biol Reprod. 2013 Feb 14;88(2):35. doi: 10.1095/biolreprod.112.105601. Print 2013 Feb. Biol Reprod. 2013. PMID: 23197164 Review.
Cited by
-
Review on Genomic Regions and Candidate Genes Associated with Economically Important Production and Reproduction Traits in Sheep (Ovies aries).Animals (Basel). 2019 Dec 23;10(1):33. doi: 10.3390/ani10010033. Animals (Basel). 2019. PMID: 31877963 Free PMC article. Review.
-
Overexpression of Lhx8 inhibits cell proliferation and induces cell cycle arrest in PC12 cell line.In Vitro Cell Dev Biol Anim. 2015 Apr;51(4):329-35. doi: 10.1007/s11626-014-9838-y. Epub 2014 Dec 5. In Vitro Cell Dev Biol Anim. 2015. PMID: 25475040
-
Recent Advances in the Understanding of Gonadotrope Lineage Differentiation in the Developing Pituitary.Neuroendocrinology. 2025;115(2):195-210. doi: 10.1159/000542513. Epub 2024 Nov 11. Neuroendocrinology. 2025. PMID: 39527929 Free PMC article. Review.
-
Physiological and Molecular Mechanisms of Differential Sensitivity of Palmer Amaranth (Amaranthus palmeri) to Mesotrione at Varying Growth Temperatures.PLoS One. 2015 May 19;10(5):e0126731. doi: 10.1371/journal.pone.0126731. eCollection 2015. PLoS One. 2015. PMID: 25992558 Free PMC article.
-
Gene targeting of mouse Tardbp negatively affects Masp2 expression.PLoS One. 2014 Apr 16;9(4):e95373. doi: 10.1371/journal.pone.0095373. eCollection 2014. PLoS One. 2014. PMID: 24740308 Free PMC article.
References
-
- Mollard P, Hodson DJ, Lafont C, Rizzoti K, Drouin J. A tridimensional view of pituitary development and function. Trends Endocrinol Metab. 2012;23:261–269 - PubMed
-
- Prince KL, Walvoord EC, Rhodes SJ. The role of homeodomain transcription factors in heritable pituitary disease. Nat Rev Endocrinol. 2011;7:727–737 - PubMed
-
- Simmons DM, Voss JW, Ingraham HA, et al. Pituitary cell phenotypes involve cell-specific Pit-1 mRNA translation and synergistic interactions with other classes of transcription factors. Genes Dev. 1990;4:695–711 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

