Pleiotropic constraints, expression level, and the evolution of miRNA sequences
- PMID: 24100521
- PMCID: PMC3913099
- DOI: 10.1007/s00239-013-9588-6
Pleiotropic constraints, expression level, and the evolution of miRNA sequences
Abstract
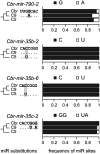
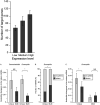
Post-transcriptional gene regulation mediated by microRNAs (miRNAs) plays critical roles during development by modulating gene expression and conferring robustness to stochastic errors. Phylogenetic analyses suggest that miRNA acquisition could play a role in phenotypic innovation. Moreover, miRNA-induced regulation strongly impacts genome evolution, increasing selective constraints on 3'UTRs, protein sequences, and expression level divergence. Thus, it is essential to understand the factors governing sequence evolution for this important class of regulatory molecules. Investigation of the patterns of molecular evolution at miRNA loci have been limited in Caenorhabditis elegans because of the lack of a close outgroup. Instead, I used Caenorhabditis briggsae as the focus point of this study because of its close relationship to Caenorhabditis sp. 9. I also corroborated the patterns of sequence evolution in Caenorhabditis using published orthologous relationships among miRNAs in Drosophila. In nematodes and in flies, miRNA sequence divergence is not influenced by the genomic neighborhood (i.e., intronic or intergenic) but is nevertheless affected by the genomic context because X-linked miRNAs evolve faster than autosomal miRNAs. However, this effect of chromosomal linkage can be explained by differential expression levels rather than a fast-X effect. The results presented here support a universal negative relationship between rates of molecular evolution and expression level, and suggest that mutations in highly expressed miRNAs are more likely to be deleterious because they potentially affect a larger number of target genes. Finally, I show that many single family member miRNAs evolve faster than miRNAs from multigene families and have limited functional scope, suggesting that they are not strongly integrated in gene regulatory networks.
Figures

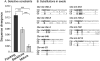

Similar articles
-
Repertoire and evolution of miRNA genes in four divergent nematode species.Genome Res. 2009 Nov;19(11):2064-74. doi: 10.1101/gr.093781.109. Epub 2009 Sep 15. Genome Res. 2009. PMID: 19755563 Free PMC article.
-
Functional constraint and divergence in the G protein family in Caenorhabditis elegans and Caenorhabditis briggsae.Mol Genet Genomics. 2005 Jun;273(4):299-310. doi: 10.1007/s00438-004-1105-6. Epub 2005 Apr 27. Mol Genet Genomics. 2005. PMID: 15856303
-
Adaptive evolution of testis-specific, recently evolved, clustered miRNAs in Drosophila.RNA. 2014 Aug;20(8):1195-209. doi: 10.1261/rna.044644.114. Epub 2014 Jun 18. RNA. 2014. PMID: 24942624 Free PMC article.
-
Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny.DNA Cell Biol. 2007 Apr;26(4):209-18. doi: 10.1089/dna.2006.0545. DNA Cell Biol. 2007. PMID: 17465887 Review.
-
Evolution of microRNA diversity and regulation in animals.Nat Rev Genet. 2011 Nov 18;12(12):846-60. doi: 10.1038/nrg3079. Nat Rev Genet. 2011. PMID: 22094948 Review.
Cited by
-
Comparative genomic analysis of upstream miRNA regulatory motifs in Caenorhabditis.RNA. 2016 Jul;22(7):968-78. doi: 10.1261/rna.055392.115. Epub 2016 May 2. RNA. 2016. PMID: 27140965 Free PMC article.
-
Parallel faster-X evolution of gene expression and protein sequences in Drosophila: beyond differences in expression properties and protein interactions.PLoS One. 2015 Mar 19;10(3):e0116829. doi: 10.1371/journal.pone.0116829. eCollection 2015. PLoS One. 2015. PMID: 25789611 Free PMC article.
-
A suppression-modification gene drive for malaria control targeting the ultra-conserved RNA gene mir-184.Nat Commun. 2025 Apr 25;16(1):3923. doi: 10.1038/s41467-025-58954-5. Nat Commun. 2025. PMID: 40280899 Free PMC article.
-
Microevolution of nematode miRNAs reveals diverse modes of selection.Genome Biol Evol. 2014 Oct 28;6(11):3049-63. doi: 10.1093/gbe/evu239. Genome Biol Evol. 2014. PMID: 25355809 Free PMC article.
References
-
- Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–93. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

