Population genomic footprints of selection and associations with climate in natural populations of Arabidopsis halleri from the Alps
- PMID: 24102711
- PMCID: PMC4274019
- DOI: 10.1111/mec.12521
Population genomic footprints of selection and associations with climate in natural populations of Arabidopsis halleri from the Alps
Abstract
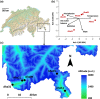
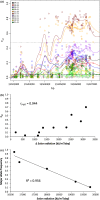
Natural genetic variation is essential for the adaptation of organisms to their local environment and to changing environmental conditions. Here, we examine genomewide patterns of nucleotide variation in natural populations of the outcrossing herb Arabidopsis halleri and associations with climatic variation among populations in the Alps. Using a pooled population sequencing (Pool-Seq) approach, we discovered more than two million SNPs in five natural populations and identified highly differentiated genomic regions and SNPs using FST -based analyses. We tested only the most strongly differentiated SNPs for associations with a nonredundant set of environmental factors using partial Mantel tests to identify topo-climatic factors that may underlie the observed footprints of selection. Possible functions of genes showing signatures of selection were identified by Gene Ontology analysis. We found 175 genes to be highly associated with one or more of the five tested topo-climatic factors. Of these, 23.4% had unknown functions. Genetic variation in four candidate genes was strongly associated with site water balance and solar radiation, and functional annotations were congruent with these environmental factors. Our results provide a genomewide perspective on the distribution of adaptive genetic variation in natural plant populations from a highly diverse and heterogeneous alpine environment.
Keywords: Pool-Seq; adaptation; environmental association; genome resequencing; pooled sequencing; population genomics.
© 2013 John Wiley & Sons Ltd.
Figures


Similar articles
-
Local adaptation (mostly) remains local: reassessing environmental associations of climate-related candidate SNPs in Arabidopsis halleri.Heredity (Edinb). 2017 Feb;118(2):193-201. doi: 10.1038/hdy.2016.82. Epub 2016 Oct 5. Heredity (Edinb). 2017. PMID: 27703154 Free PMC article.
-
Back to nature: ecological genomics of loblolly pine (Pinus taeda, Pinaceae).Mol Ecol. 2010 Sep;19(17):3789-805. doi: 10.1111/j.1365-294X.2010.04698.x. Epub 2010 Aug 13. Mol Ecol. 2010. PMID: 20723060
-
The genomic footprint of climate adaptation in Chironomus riparius.Mol Ecol. 2018 Mar;27(6):1439-1456. doi: 10.1111/mec.14543. Epub 2018 Mar 23. Mol Ecol. 2018. PMID: 29473242
-
Genetic and genomic approaches to assess adaptive genetic variation in plants: forest trees as a model.Physiol Plant. 2009 Dec;137(4):509-19. doi: 10.1111/j.1399-3054.2009.01263.x. Epub 2009 Jun 12. Physiol Plant. 2009. PMID: 19627554 Review.
-
Controlling false discoveries in genome scans for selection.Mol Ecol. 2016 Jan;25(2):454-69. doi: 10.1111/mec.13513. Epub 2016 Jan 18. Mol Ecol. 2016. PMID: 26671840 Review.
Cited by
-
On the Causes of Rapid Diversification in the Páramos: Isolation by Ecology and Genomic Divergence in Espeletia.Front Plant Sci. 2018 Dec 3;9:1700. doi: 10.3389/fpls.2018.01700. eCollection 2018. Front Plant Sci. 2018. PMID: 30581444 Free PMC article.
-
Tropical forest loss and geographic location drive the functional genomic diversity of an endangered palm tree.Evol Appl. 2023 Jun 15;16(7):1257-1273. doi: 10.1111/eva.13525. eCollection 2023 Jul. Evol Appl. 2023. PMID: 37492151 Free PMC article.
-
Continental-scale footprint of balancing and positive selection in a small rodent (Microtus arvalis).PLoS One. 2014 Nov 10;9(11):e112332. doi: 10.1371/journal.pone.0112332. eCollection 2014. PLoS One. 2014. PMID: 25383542 Free PMC article.
-
Integrating very high resolution environmental proxies in genotype-environment association studies.Evol Appl. 2024 Jun 28;17(7):e13737. doi: 10.1111/eva.13737. eCollection 2024 Jul. Evol Appl. 2024. PMID: 38948540 Free PMC article.
-
A Comprehensive Review on Chickpea (Cicer arietinum L.) Breeding for Abiotic Stress Tolerance and Climate Change Resilience.Int J Mol Sci. 2022 Jun 18;23(12):6794. doi: 10.3390/ijms23126794. Int J Mol Sci. 2022. PMID: 35743237 Free PMC article. Review.
References
-
- Alexa A, Rahnenührer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. - PubMed
-
- Andrew RL, Bernatchez L, Bonin A, et al. A road map for molecular ecology. Molecular Ecology. 2013;22:2605–2626. - PubMed
-
- Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends in Ecology and Evolution. 2008;23:38–44. - PubMed
-
- Bergelson J, Roux F. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nature Reviews Genetics. 2010;11:867–879. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

