Canonical transforming growth factor-β signaling regulates disintegrin metalloprotease expression in experimental renal fibrosis via miR-29
- PMID: 24103556
- PMCID: PMC4188136
- DOI: 10.1016/j.ajpath.2013.08.027
Canonical transforming growth factor-β signaling regulates disintegrin metalloprotease expression in experimental renal fibrosis via miR-29
Abstract
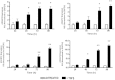
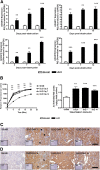
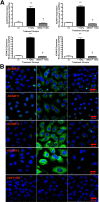
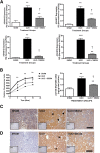
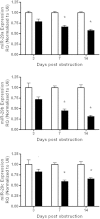
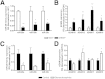
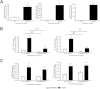
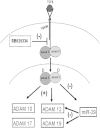
Fibrosis pathophysiology is critically regulated by Smad 2- and Smad 3-mediated transforming growth factor-β (TGF-β) signaling. Disintegrin metalloproteases (Adam) can manipulate the signaling environment, however, the role and regulation of ADAMs in renal fibrosis remain unclear. TGF-β stimulation of renal cells results in a significant up-regulation of Adams 10, 17, 12, and 19. The selective Smad2/3 inhibitor SB 525334 reversed these TGF-β-induced changes. In vivo, using ureteral obstruction to model renal fibrosis, we observed increased Adams gene expression that was blocked by oral administration of SB 525334. Similar increases in Adam gene expression also occurred in preclinical models of hypertension-induced renal damage and glomerulonephritis. miRNAs are a recently discovered second level of regulation of gene expression. Analysis of 3' untranslated regions of Adam12 and Adam19 mRNAs showed multiple binding sites for miR-29a, miR-29b, and miR-29c. We show that miR-29 family expression is decreased after unilateral ureter obstruction and this significant decrease in miR-29 family expression was observed consistently in preclinical models of renal dysfunction and correlated with an increase in Adam12 and Adam19 expression. Exogenous overexpression of the miR-29 family blocked TGF-β-mediated up-regulation of Adam12 and Adam19 gene expression. This study shows that Adams are involved in renal fibrosis and are regulated by canonical TGF-β signaling and miR-29. Therefore, both Adams and the miR-29 family represent therapeutic targets for renal fibrosis.
Copyright © 2013 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Bohle A., Mackensen-Haen S., von Gise H. Significance of tubulointerstitial changes in the renal cortex for the excretory function and concentration ability of the kidney: a morphometric contribution. Am J Nephrol. 1987;7:421–433. - PubMed
-
- Bottinger E.P. TGF-beta in renal injury and disease. Semin Nephrol. 2007;27:309–320. - PubMed
-
- Eddy A.A., Neilson E.G. Chronic kidney disease progression. J Am Soc Nephrol. 2006;17:2964–2966. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

