Reticulate evolution of the rye genome
- PMID: 24104565
- PMCID: PMC3877785
- DOI: 10.1105/tpc.113.114553
Reticulate evolution of the rye genome
Abstract
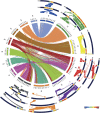
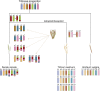
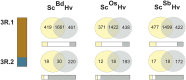
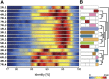
Rye (Secale cereale) is closely related to wheat (Triticum aestivum) and barley (Hordeum vulgare). Due to its large genome (~8 Gb) and its regional importance, genome analysis of rye has lagged behind other cereals. Here, we established a virtual linear gene order model (genome zipper) comprising 22,426 or 72% of the detected set of 31,008 rye genes. This was achieved by high-throughput transcript mapping, chromosome survey sequencing, and integration of conserved synteny information of three sequenced model grass genomes (Brachypodium distachyon, rice [Oryza sativa], and sorghum [Sorghum bicolor]). This enabled a genome-wide high-density comparative analysis of rye/barley/model grass genome synteny. Seventeen conserved syntenic linkage blocks making up the rye and barley genomes were defined in comparison to model grass genomes. Six major translocations shaped the modern rye genome in comparison to a putative Triticeae ancestral genome. Strikingly dissimilar conserved syntenic gene content, gene sequence diversity signatures, and phylogenetic networks were found for individual rye syntenic blocks. This indicates that introgressive hybridizations (diploid or polyploidy hybrid speciation) and/or a series of whole-genome or chromosome duplications played a role in rye speciation and genome evolution.
Figures

References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

