A 7-deoxyloganetic acid glucosyltransferase contributes a key step in secologanin biosynthesis in Madagascar periwinkle
- PMID: 24104568
- PMCID: PMC3877786
- DOI: 10.1105/tpc.113.115154
A 7-deoxyloganetic acid glucosyltransferase contributes a key step in secologanin biosynthesis in Madagascar periwinkle
Abstract
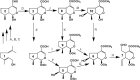
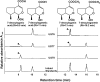
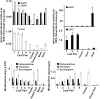
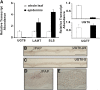
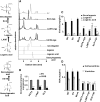
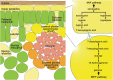
Iridoids form a broad and versatile class of biologically active molecules found in thousands of plant species. In addition to the many hundreds of iridoids occurring in plants, some iridoids, such as secologanin, serve as key building blocks in the biosynthesis of thousands of monoterpene indole alkaloids (MIAs) and many quinoline alkaloids. This study describes the molecular cloning and functional characterization of three iridoid glucosyltransfeases (UDP-sugar glycosyltransferase6 [UGT6], UGT7, and UGT8) from Madagascar periwinkle (Catharanthus roseus) with remarkably different catalytic efficiencies. Biochemical analyses reveal that UGT8 possessed a high catalytic efficiency toward its exclusive iridoid substrate, 7-deoxyloganetic acid, making it better suited for the biosynthesis of iridoids in periwinkle than the other two iridoid glucosyltransfeases. The role of UGT8 in the fourth to last step in secologanin biosynthesis was confirmed by virus-induced gene silencing in periwinkle plants, which reduced expression of this gene and resulted in a large decline in secologanin and MIA accumulation within silenced plants. Localization studies of UGT8 using a carborundum abrasion method for RNA extraction show that its expression occurs preferentially within periwinkle leaves rather than in epidermal cells, and in situ hybridization studies confirm that UGT8 is preferentially expressed in internal phloem associated parenchyma cells of periwinkle species.
Figures
References
-
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 - PubMed
-
- Burlat V., Oudin A., Courtois M., Rideau M., St-Pierre B. (2004). Co-expression of three MEP pathway genes and geraniol 10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J. 38: 131–141 - PubMed
-
- De Luca V., Salim V., Atsumi S.M., Yu F. (2012a). Mining the biodiversity of plants: A revolution in the making. Science 336: 1658–1661 - PubMed
-
- De Luca V., Salim V., Levac D., Atsumi S.M., Yu F. (2012b). Discovery and functional analysis of monoterpenoid indole alkaloid pathways in plants. Methods Enzymol. 515: 207–229 - PubMed
-
- Facchini P.J., Bohlmann J., Covello P.S., De Luca V., Mahadevan R., Page J.E., Ro D.K., Sensen C.W., Storms R., Martin V.J. (2012). Synthetic biosystems for the production of high-value plant metabolites. Trends Biotechnol. 30: 127–131 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

