Morphine biosynthesis in opium poppy involves two cell types: sieve elements and laticifers
- PMID: 24104569
- PMCID: PMC3877807
- DOI: 10.1105/tpc.113.115113
Morphine biosynthesis in opium poppy involves two cell types: sieve elements and laticifers
Abstract
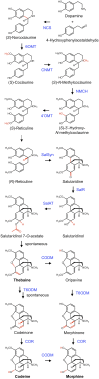
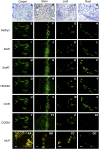
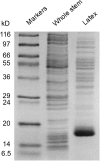
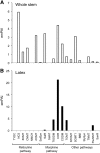
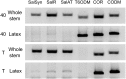
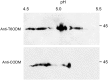
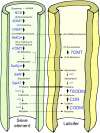
Immunofluorescence labeling and shotgun proteomics were used to establish the cell type-specific localization of morphine biosynthesis in opium poppy (Papaver somniferum). Polyclonal antibodies for each of six enzymes involved in converting (R)-reticuline to morphine detected corresponding antigens in sieve elements of the phloem, as described previously for all upstream enzymes transforming (S)-norcoclaurine to (S)-reticuline. Validated shotgun proteomics performed on whole-stem and latex total protein extracts generated 2031 and 830 distinct protein families, respectively. Proteins corresponding to nine morphine biosynthetic enzymes were represented in the whole stem, whereas only four of the final five pathway enzymes were detected in the latex. Salutaridine synthase was detected in the whole stem, but not in the latex subproteome. The final three enzymes converting thebaine to morphine were among the most abundant active latex proteins despite a limited occurrence in laticifers suggested by immunofluorescence labeling. Multiple charge isoforms of two key O-demethylases in the latex were revealed by two-dimensional immunoblot analysis. Salutaridine biosynthesis appears to occur only in sieve elements, whereas conversion of thebaine to morphine is predominant in adjacent laticifers, which contain morphine-rich latex. Complementary use of immunofluorescence labeling and shotgun proteomics has substantially resolved the cellular localization of morphine biosynthesis in opium poppy.
Figures

References
-
- Brochmann-Hanssen E. (1984). A second pathway for the terminal steps in the biosynthesis of morphine. Planta Med. 50: 343–345 - PubMed
-
- Cho W.K., Chen X.Y., Rim Y., Chu H., Jo Y., Kim S., Park Z.-Y., Kim J.Y. (2010). Extended latex proteome analysis deciphers additional roles of the lettuce laticifer. Plant Biotechnol. Rep. 4: 311–319
-
- D’Amato A., Bachi A., Fasoli E., Boschetti E., Peltre G., Sénéchal H., Sutra J.P., Citterio A., Righetti P.G. (2010). In-depth exploration of Hevea brasiliensis latex proteome and “hidden allergens” via combinatorial peptide ligand libraries. J. Proteomics 73: 1368–1380 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

