Regions outside the DNA-binding domain are critical for proper in vivo specificity of an archetypal zinc finger transcription factor
- PMID: 24106088
- PMCID: PMC3874204
- DOI: 10.1093/nar/gkt895
Regions outside the DNA-binding domain are critical for proper in vivo specificity of an archetypal zinc finger transcription factor
Abstract
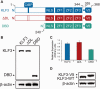
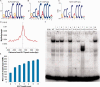
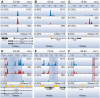
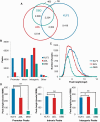
Transcription factors (TFs) are often regarded as being composed of a DNA-binding domain (DBD) and a functional domain. The two domains are considered separable and autonomous, with the DBD directing the factor to its target genes and the functional domain imparting transcriptional regulation. We examined an archetypal zinc finger (ZF) TF, Krüppel-like factor 3 with an N-terminal domain that binds the corepressor CtBP and a DBD composed of three ZFs at its C-terminus. We established a system to compare the genomic occupancy profile of wild-type Krüppel-like factor 3 with two mutants affecting the N-terminal functional domain: a mutant unable to contact the cofactor CtBP and a mutant lacking the entire N-terminal domain, but retaining the ZFs intact. Chromatin immunoprecipitation followed by sequencing was used to assess binding across the genome in murine embryonic fibroblasts. Unexpectedly, we observe that mutations in the N-terminal domain generally reduced binding, but there were also instances where binding was retained or even increased. These results provide a clear demonstration that the correct localization of TFs to their target genes is not solely dependent on their DNA-contact domains. This informs our understanding of how TFs operate and is of relevance to the design of artificial ZF proteins.
Figures


References
-
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. - PubMed
-
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

