Structures of the substrate-free and product-bound forms of HmuO, a heme oxygenase from corynebacterium diphtheriae: x-ray crystallography and molecular dynamics investigation
- PMID: 24106279
- PMCID: PMC3843059
- DOI: 10.1074/jbc.M113.486936
Structures of the substrate-free and product-bound forms of HmuO, a heme oxygenase from corynebacterium diphtheriae: x-ray crystallography and molecular dynamics investigation
Abstract
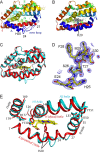
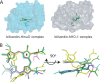
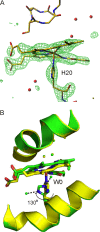
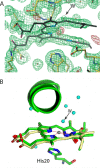
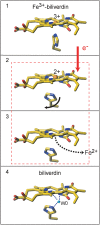
Heme oxygenase catalyzes the degradation of heme to biliverdin, iron, and carbon monoxide. Here, we present crystal structures of the substrate-free, Fe(3+)-biliverdin-bound, and biliverdin-bound forms of HmuO, a heme oxygenase from Corynebacterium diphtheriae, refined to 1.80, 1.90, and 1.85 Å resolution, respectively. In the substrate-free structure, the proximal and distal helices, which tightly bracket the substrate heme in the substrate-bound heme complex, move apart, and the proximal helix is partially unwound. These features are supported by the molecular dynamic simulations. The structure implies that the heme binding fixes the enzyme active site structure, including the water hydrogen bond network critical for heme degradation. The biliverdin groups assume the helical conformation and are located in the heme pocket in the crystal structures of the Fe(3+)-biliverdin-bound and the biliverdin-bound HmuO, prepared by in situ heme oxygenase reaction from the heme complex crystals. The proximal His serves as the Fe(3+)-biliverdin axial ligand in the former complex and forms a hydrogen bond through a bridging water molecule with the biliverdin pyrrole nitrogen atoms in the latter complex. In both structures, salt bridges between one of the biliverdin propionate groups and the Arg and Lys residues further stabilize biliverdin at the HmuO heme pocket. Additionally, the crystal structure of a mixture of two intermediates between the Fe(3+)-biliverdin and biliverdin complexes has been determined at 1.70 Å resolution, implying a possible route for iron exit.
Keywords: Bacterial Iron Acquisition; Enzyme Structure; Heme; Heme Oxygenase; Molecular Dynamics; X-ray Crystallography.
Figures




References
-
- Maines M. D. (1997) The heme oxygenase system: a regulator of second messenger gases. Annu. Rev. Pharmacol. Toxicol. 37, 517–554 - PubMed
-
- Schacter B. A., Nelson E. B., Marver H. S., Masters B. S. (1972) Immunochemical evidence for an association of heme oxygenase with the microsomal electron transport system. J. Biol. Chem. 247, 3601–3607 - PubMed
-
- Boehning D., Snyder S. H. (2003) Novel neural modulators. Annu. Rev. Neurosci. 26, 105–131 - PubMed
-
- Paul B. D., Snyder S. H. (2012) H2S signaling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 13, 499–507 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

