Meiotic chromosome structures constrain and respond to designation of crossover sites
- PMID: 24107990
- PMCID: PMC3920622
- DOI: 10.1038/nature12577
Meiotic chromosome structures constrain and respond to designation of crossover sites
Abstract
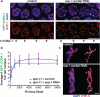
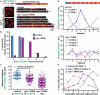
Crossover recombination events between homologous chromosomes are required to form chiasmata, temporary connections between homologues that ensure their proper segregation at meiosis I. Despite this requirement for crossovers and an excess of the double-strand DNA breaks that are the initiating events for meiotic recombination, most organisms make very few crossovers per chromosome pair. Moreover, crossovers tend to inhibit the formation of other crossovers nearby on the same chromosome pair, a poorly understood phenomenon known as crossover interference. Here we show that the synaptonemal complex, a meiosis-specific structure that assembles between aligned homologous chromosomes, both constrains and is altered by crossover recombination events. Using a cytological marker of crossover sites in Caenorhabditis elegans, we show that partial depletion of the synaptonemal complex central region proteins attenuates crossover interference, increasing crossovers and reducing the effective distance over which interference operates, indicating that synaptonemal complex proteins limit crossovers. Moreover, we show that crossovers are associated with a local 0.4-0.5-micrometre increase in chromosome axis length. We propose that meiotic crossover regulation operates as a self-limiting system in which meiotic chromosome structures establish an environment that promotes crossover formation, which in turn alters chromosome structure to inhibit other crossovers at additional sites.
Figures
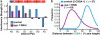

Comment in
-
Meiosis: SYP up firmly and cross over evenly.Curr Biol. 2014 Jan 6;24(1):R20-R23. doi: 10.1016/j.cub.2013.11.007. Curr Biol. 2014. PMID: 24405671
References
-
- Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–789. - PubMed
-
- Muller HJ. The mechanism of crossing-over. Am. Naturalist. 1916;50:193–221.
-
- Sturtevant AH. The linear arrangements of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Zoology. 1913;14:43–59.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

