Gene-targeted metagenomic analysis of glucan-branching enzyme gene profiles among human and animal fecal microbiota
- PMID: 24108330
- PMCID: PMC3930310
- DOI: 10.1038/ismej.2013.167
Gene-targeted metagenomic analysis of glucan-branching enzyme gene profiles among human and animal fecal microbiota
Abstract
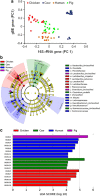
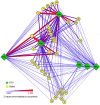
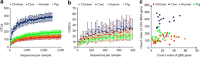
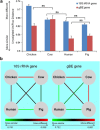
Glycoside hydrolases (GHs), the enzymes that breakdown complex carbohydrates, are a highly diversified class of key enzymes associated with the gut microbiota and its metabolic functions. To learn more about the diversity of GHs and their potential role in a variety of gut microbiomes, we used a combination of 16S, metagenomic and targeted amplicon sequencing data to study one of these enzyme families in detail. Specifically, we employed a functional gene-targeted metagenomic approach to the 1-4-α-glucan-branching enzyme (gBE) gene in the gut microbiomes of four host species (human, chicken, cow and pig). The characteristics of operational taxonomic units (OTUs) and operational glucan-branching units (OGBUs) were distinctive in each of hosts. Human and pig were most similar in OTUs profiles while maintaining distinct OGBU profiles. Interestingly, the phylogenetic profiles identified from 16S and gBE gene sequences differed, suggesting the presence of different gBE genes in the same OTU across different vertebrate hosts. Our data suggest that gene-targeted metagenomic analysis is useful for an in-depth understanding of the diversity of a particular gene of interest. Specific carbohydrate metabolic genes appear to be carried by distinct OTUs in different individual hosts and among different vertebrate species' microbiomes, the characteristics of which differ according to host genetic background and/or diet.
Figures

References
-
- Callaway T, Dowd S, Edrington T, Anderson R, Krueger N, Bauer N, et al. Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J Anim Sci. 2010;88:3977–3983. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

