Next-generation sequence analysis of the genome of RFHVMn, the macaque homolog of Kaposi's sarcoma (KS)-associated herpesvirus, from a KS-like tumor of a pig-tailed macaque
- PMID: 24109218
- PMCID: PMC3838293
- DOI: 10.1128/JVI.02331-13
Next-generation sequence analysis of the genome of RFHVMn, the macaque homolog of Kaposi's sarcoma (KS)-associated herpesvirus, from a KS-like tumor of a pig-tailed macaque
Abstract
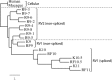
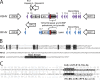
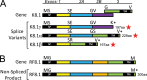
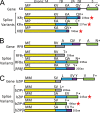
The complete sequence of retroperitoneal fibromatosis-associated herpesvirus Macaca nemestrina (RFHVMn), the pig-tailed macaque homolog of Kaposi's sarcoma-associated herpesvirus (KSHV), was determined by next-generation sequence analysis of a Kaposi's sarcoma (KS)-like macaque tumor. Colinearity of genes was observed with the KSHV genome, and the core herpesvirus genes had strong sequence homology to the corresponding KSHV genes. RFHVMn lacked homologs of open reading frame 11 (ORF11) and KSHV ORFs K5 and K6, which appear to have been generated by duplication of ORFs K3 and K4 after the divergence of KSHV and RFHV. RFHVMn contained positional homologs of all other unique KSHV genes, although some showed limited sequence similarity. RFHVMn contained a number of candidate microRNA genes. Although there was little sequence similarity with KSHV microRNAs, one candidate contained the same seed sequence as the positional homolog, kshv-miR-K12-10a, suggesting functional overlap. RNA transcript splicing was highly conserved between RFHVMn and KSHV, and strong sequence conservation was noted in specific promoters and putative origins of replication, predicting important functional similarities. Sequence comparisons indicated that RFHVMn and KSHV developed in long-term synchrony with the evolution of their hosts, and both viruses phylogenetically group within the RV1 lineage of Old World primate rhadinoviruses. RFHVMn is the closest homolog of KSHV to be completely sequenced and the first sequenced RV1 rhadinovirus homolog of KSHV from a nonhuman Old World primate. The strong genetic and sequence similarity between RFHVMn and KSHV, coupled with similarities in biology and pathology, demonstrate that RFHVMn infection in macaques offers an important and relevant model for the study of KSHV in humans.
Figures








Similar articles
-
Complete genome sequence of Pig-tailed macaque rhadinovirus 2 and its evolutionary relationship with rhesus macaque rhadinovirus and human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus.J Virol. 2015 Apr;89(7):3888-909. doi: 10.1128/JVI.03597-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609822 Free PMC article.
-
Analysis of 4.3 kilobases of divergent locus B of macaque retroperitoneal fibromatosis-associated herpesvirus reveals a close similarity in gene sequence and genome organization to Kaposi's sarcoma-associated herpesvirus.J Virol. 2003 May;77(9):5084-97. doi: 10.1128/jvi.77.9.5084-5097.2003. J Virol. 2003. PMID: 12692211 Free PMC article.
-
Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus.J Virol. 2000 May;74(10):4919-28. doi: 10.1128/jvi.74.10.4919-4928.2000. J Virol. 2000. PMID: 10775636 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus and Kaposi's sarcoma.Microbes Infect. 2000 May;2(6):671-80. doi: 10.1016/s1286-4579(00)00358-0. Microbes Infect. 2000. PMID: 10884618 Review.
-
Comparative pathobiology of Kaposi sarcoma-associated herpesvirus and related primate rhadinoviruses.Comp Med. 2008 Feb;58(1):31-42. Comp Med. 2008. PMID: 19793454 Free PMC article. Review.
Cited by
-
STAT and Janus kinase targeting by human herpesvirus 8 interferon regulatory factor in the suppression of type-I interferon signaling.PLoS Pathog. 2022 Jul 1;18(7):e1010676. doi: 10.1371/journal.ppat.1010676. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35776779 Free PMC article.
-
ORF73 LANA homologs of RRV and MneRV2 contain an extended RGG/RG-rich nuclear and nucleolar localization signal that interacts directly with importin β1 for non-classical nuclear import.Virology. 2017 Nov;511:152-164. doi: 10.1016/j.virol.2017.08.029. Epub 2017 Aug 29. Virology. 2017. PMID: 28850829 Free PMC article.
-
Novel Virus Related to Kaposi's Sarcoma-Associated Herpesvirus from Colobus Monkey.Emerg Infect Dis. 2019 Aug;25(8):1548-1551. doi: 10.3201/eid2508.181802. Emerg Infect Dis. 2019. PMID: 31310220 Free PMC article.
-
Fetal brain lesions after subcutaneous inoculation of Zika virus in a pregnant nonhuman primate.Nat Med. 2016 Nov;22(11):1256-1259. doi: 10.1038/nm.4193. Epub 2016 Sep 12. Nat Med. 2016. PMID: 27618651 Free PMC article.
-
Experimental co-transmission of Simian Immunodeficiency Virus (SIV) and the macaque homologs of the Kaposi Sarcoma-Associated Herpesvirus (KSHV) and Epstein-Barr Virus (EBV).PLoS One. 2018 Nov 16;13(11):e0205632. doi: 10.1371/journal.pone.0205632. eCollection 2018. PLoS One. 2018. PMID: 30444879 Free PMC article.
References
-
- Antman K, Chang Y. 2000. Kaposi's sarcoma. N. Engl. J. Med. 342:1027–1038 - PubMed
-
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869 - PubMed
-
- Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande JP, Weiss RA, Alitalo K, Boshoff C. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. U. S. A. 96:4546–4551 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

