Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology
- PMID: 24109552
- PMCID: PMC3792188
- DOI: 10.7717/peerj.167
Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology
Abstract
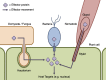
The Galaxy Project offers the popular web browser-based platform Galaxy for running bioinformatics tools and constructing simple workflows. Here, we present a broad collection of additional Galaxy tools for large scale analysis of gene and protein sequences. The motivating research theme is the identification of specific genes of interest in a range of non-model organisms, and our central example is the identification and prediction of "effector" proteins produced by plant pathogens in order to manipulate their host plant. This functional annotation of a pathogen's predicted capacity for virulence is a key step in translating sequence data into potential applications in plant pathology. This collection includes novel tools, and widely-used third-party tools such as NCBI BLAST+ wrapped for use within Galaxy. Individual bioinformatics software tools are typically available separately as standalone packages, or in online browser-based form. The Galaxy framework enables the user to combine these and other tools to automate organism scale analyses as workflows, without demanding familiarity with command line tools and scripting. Workflows created using Galaxy can be saved and are reusable, so may be distributed within and between research groups, facilitating the construction of a set of standardised, reusable bioinformatic protocols. The Galaxy tools and workflows described in this manuscript are open source and freely available from the Galaxy Tool Shed (http://usegalaxy.org/toolshed or http://toolshed.g2.bx.psu.edu).
Keywords: Accessibility; Annotation; Effector proteins; Galaxy; Genomics; Pipeline; Reproducibility; Sequence analysis; Workflow.
Figures
References
-
- Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathogens. 2011;7(7):e1002132. doi: 10.1371/journal.ppat.1002132. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

