Down-modulation of nucleoporin RanBP2/Nup358 impaired chromosomal alignment and induced mitotic catastrophe
- PMID: 24113188
- PMCID: PMC3824679
- DOI: 10.1038/cddis.2013.370
Down-modulation of nucleoporin RanBP2/Nup358 impaired chromosomal alignment and induced mitotic catastrophe
Abstract
Chromosomal missegregation is a common feature of many human tumors. Recent studies have indicated a link between nucleoporin RanBP2/Nup358 and chromosomal segregation during mitosis; however, the molecular details have yet to be fully established. Observed through live cell imaging and flow cytometry, here we show that RNA interference-mediated knockdown of RanBP2 induced G2/M phase arrest, metaphase catastrophe and mitotic cell death. Furthermore, RanBP2 down-modulation disrupted importin/karyopherin β1 as well as the expression and localization of the Ran GTPase activating protein 1. We found that N-terminal of RanBP2 interacted with the N-terminal of importin β1. Moreover, at least a portion of RanBP2 partially localizes at the centrosome during mitosis. Notably, we also found that GTPase Ran is also involved in the regulation of RanBP2-importin β1 interaction. Overall, our results suggest that mitotic arrest and the following cell death were caused by depletion of RanBP2. Our findings point to a crucial role for RanBP2 in proper mitotic progression and faithful chromosomal segregation.
Figures
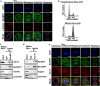

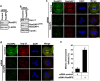




References
-
- Nakano H, Wang W, Hashizume C, Funasaka T, Sato H, Wong RW. Unexpected role of nucleoporins in coordination of cell cycle progression. Cell Cycle. 2011;10:425–433. - PubMed
-
- Guttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. - PubMed
-
- Raices M, D'Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13:687–699. - PubMed
-
- Bukata L, Parker SL, D'Angelo MA. Nuclear pore complexes in the maintenance of genome integrity. Curr Opin Cell Biol. 2013;25:378–386. - PubMed
-
- Wong RW. Interaction between Rae1 and cohesin subunit SMC1 is required for proper spindle formation. Cell Cycle. 2010;9:198–200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

