No need for a power stroke in ISWI-mediated nucleosome sliding
- PMID: 24113208
- PMCID: PMC3981079
- DOI: 10.1038/embor.2013.160
No need for a power stroke in ISWI-mediated nucleosome sliding
Abstract
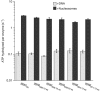
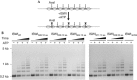
Nucleosome remodelling enzymes of the ISWI family reposition nucleosomes in eukaryotes. ISWI contains an ATPase and a HAND-SANT-SLIDE (HSS) domain. Conformational changes between these domains have been proposed to be critical for nucleosome repositioning by pulling flanking DNA into the nucleosome. We inserted flexible linkers at strategic sites in ISWI to disrupt this putative power stroke and assess its functional importance by quantitative biochemical assays. Notably, the flexible linkers did not disrupt catalysis. Instead of engaging in a power stroke, the HSS module might therefore assist DNA to ratchet into the nucleosome. Our results clarify the roles had by the domains and suggest that the HSS domain evolved to optimize a rudimentary remodelling engine.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes.Nature. 2012 Dec 13;492(7428):280-4. doi: 10.1038/nature11625. Epub 2012 Nov 11. Nature. 2012. PMID: 23143334 Free PMC article.
-
The ATPase domain of ISWI is an autonomous nucleosome remodeling machine.Nat Struct Mol Biol. 2013 Jan;20(1):82-9. doi: 10.1038/nsmb.2457. Epub 2012 Dec 2. Nat Struct Mol Biol. 2013. PMID: 23202585
-
ISWI chromatin remodeling: one primary actor or a coordinated effort?Curr Opin Struct Biol. 2014 Feb;24:150-5. doi: 10.1016/j.sbi.2014.01.010. Epub 2014 Feb 19. Curr Opin Struct Biol. 2014. PMID: 24561830 Free PMC article. Review.
-
Nucleosome spacing generated by ISWI and CHD1 remodelers is constant regardless of nucleosome density.Mol Cell Biol. 2015 May;35(9):1588-605. doi: 10.1128/MCB.01070-14. Epub 2015 Mar 2. Mol Cell Biol. 2015. PMID: 25733687 Free PMC article.
-
Regulation of ISWI chromatin remodelling activity.Chromosoma. 2014 Mar;123(1-2):91-102. doi: 10.1007/s00412-013-0447-4. Epub 2014 Jan 12. Chromosoma. 2014. PMID: 24414837 Review.
Cited by
-
ISWI remodelling of physiological chromatin fibres acetylated at lysine 16 of histone H4.PLoS One. 2014 Feb 6;9(2):e88411. doi: 10.1371/journal.pone.0088411. eCollection 2014. PLoS One. 2014. PMID: 24516652 Free PMC article.
-
Molecular Mechanism of Mot1, a TATA-binding Protein (TBP)-DNA Dissociating Enzyme.J Biol Chem. 2016 Jul 22;291(30):15714-26. doi: 10.1074/jbc.M116.730366. Epub 2016 Jun 2. J Biol Chem. 2016. PMID: 27255709 Free PMC article.
-
Molecular basis of chromatin remodeling by Rhp26, a yeast CSB ortholog.Proc Natl Acad Sci U S A. 2019 Mar 26;116(13):6120-6129. doi: 10.1073/pnas.1818163116. Epub 2019 Mar 13. Proc Natl Acad Sci U S A. 2019. PMID: 30867290 Free PMC article.
-
A nucleotide-driven switch regulates flanking DNA length sensing by a dimeric chromatin remodeler.Mol Cell. 2015 Mar 5;57(5):850-859. doi: 10.1016/j.molcel.2015.01.008. Epub 2015 Feb 12. Mol Cell. 2015. PMID: 25684208 Free PMC article.
-
A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing.Nat Chem Biol. 2020 Feb;16(2):134-142. doi: 10.1038/s41589-019-0413-4. Epub 2019 Dec 9. Nat Chem Biol. 2020. PMID: 31819269 Free PMC article.
References
-
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 - PubMed
-
- Zofall M, Persinger J, Kassabov SR, Bartholomew B (2006) Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol 13: 339–346 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

