A proteolytic pathway that controls glucose uptake in fat and muscle
- PMID: 24114239
- PMCID: PMC3943767
- DOI: 10.1007/s11154-013-9276-2
A proteolytic pathway that controls glucose uptake in fat and muscle
Abstract
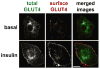
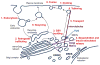
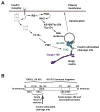
Insulin regulates glucose uptake by controlling the subcellular location of GLUT4 glucose transporters. GLUT4 is sequestered within fat and muscle cells during low-insulin states, and is translocated to the cell surface upon insulin stimulation. The TUG protein is a functional tether that sequesters GLUT4 at the Golgi matrix. To stimulate glucose uptake, insulin triggers TUG endoproteolytic cleavage. Cleavage accounts for a large proportion of the acute effect of insulin to mobilize GLUT4 to the cell surface. During ongoing insulin exposure, endocytosed GLUT4 recycles to the plasma membrane directly from endosomes, and bypasses a TUG-regulated trafficking step. Insulin acts through the TC10α GTPase and its effector protein, PIST, to stimulate TUG cleavage. This action is coordinated with insulin signals through AS160/Tbc1D4 and Tbc1D1 to modulate Rab GTPases, and with other signals to direct overall GLUT4 targeting. Data support the idea that the N-terminal TUG cleavage product, TUGUL, functions as a novel ubiquitin-like protein modifier to facilitate GLUT4 movement to the cell surface. The C-terminal TUG cleavage product is extracted from the Golgi matrix, which vacates an "anchoring" site to permit subsequent cycles of GLUT4 retention and release. Together, GLUT4 vesicle translocation and TUG cleavage may coordinate glucose uptake with physiologic effects of other proteins present in the GLUT4-containing vesicles, and with potential additional effects of the TUG C-terminal product. Understanding this TUG pathway for GLUT4 retention and release will shed light on the regulation of glucose uptake and the pathogenesis of type 2 diabetes.
Conflict of interest statement
Figures
References
-
- Cushman SW, Wardzala LJ. Potential mechanism of insulin action on glucose transport in the isolated rat adipose cell. Apparent translocation of intracellular transport systems to the plasma membrane. J Biol Chem. 1980;255(10):4758–4762. - PubMed
-
- Mueckler M, Caruso C, Baldwin SA, Panico M, Blench I, Morris HR, et al. Sequence and structure of a human glucose transporter. Science. 1985;229(4717):941–945. - PubMed
-
- Birnbaum MJ. Identification of a novel gene encoding an insulin-responsive glucose transporter protein. Cell. 1989;57(2):305–315. - PubMed
-
- James DE, Strube M, Mueckler M. Molecular cloning and characterization of an insulin-regulatable glucose transporter. Nature. 1989;338(6210):83–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

