Molecular marker differences relate to developmental position and subsets of mesodiencephalic dopaminergic neurons
- PMID: 24116087
- PMCID: PMC3792114
- DOI: 10.1371/journal.pone.0076037
Molecular marker differences relate to developmental position and subsets of mesodiencephalic dopaminergic neurons
Abstract
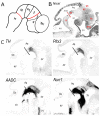
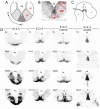
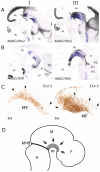
The development of mesodiencephalic dopaminergic (mdDA) neurons located in the substantia nigra compacta (SNc) and ventral tegmental area (VTA) follow a number of stages marked by distinct events. After preparation of the region by signals that provide induction and patterning, several transcription factors have been identified, which are involved in specifying the neuronal fate of these cells. The specific vulnerability of SNc neurons is thought to root in these specific developmental programs. The present study examines the positions of young postmitotic mdDA neurons to relate developmental position to mdDA subset specific markers. MdDA neurons were mapped relative to the neuromeric domains (prosomeres 1-3 (P1-3), midbrain, and hindbrain) as well as the longitudinal subdivisions (floor plate, basal plate, alar plate), as proposed by the prosomeric model. We found that postmitotic mdDA neurons are located mainly in the floorplate domain and very few in slightly more lateral domains. Moreover, mdDA neurons are present along a large proportion of the anterior/posterior axis extending from the midbrain to P3 in the diencephalon. The specific positions relate to some extent to the presence of specific subset markers as Ahd2. In the adult stage more of such subsets specific expressed genes are present and may represent a molecular map defining molecularly distinct groups of mdDA neurons.
Conflict of interest statement
Figures




References
-
- Blum D, Torch S, Lambeng N, Nissou M, Benabid AL, et al. (2001) Molecular pathways involved in the neurotoxicity of 6-OHDA, dopamine and MPTP: contribution to the apoptotic theory in Parkinson’s disease. Prog Neurobiol 65: 135–172. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

