Peptide-derivatized SB105-A10 dendrimer inhibits the infectivity of R5 and X4 HIV-1 strains in primary PBMCs and cervicovaginal histocultures
- PMID: 24116111
- PMCID: PMC3792046
- DOI: 10.1371/journal.pone.0076482
Peptide-derivatized SB105-A10 dendrimer inhibits the infectivity of R5 and X4 HIV-1 strains in primary PBMCs and cervicovaginal histocultures
Abstract
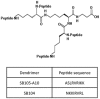
Peptide dendrimers are a class of molecules that exhibit a large array of biological effects including antiviral activity. In this report, we analyzed the antiviral activity of the peptide-derivatized SB105-A10 dendrimer, which is a tetra-branched dendrimer synthetized on a lysine core, in activated peripheral blood mononuclear cells (PBMCs) that were challenged with reference and wild-type human immunodeficiency virus type 1 (HIV-1) strains. SB105-A10 inhibited infections by HIV-1 X4 and R5 strains, interfering with the early phases of the viral replication cycle. SB105-A10 targets heparan sulfate proteoglycans (HSPGs) and, importantly, the surface plasmon resonance (SPR) assay revealed that SB105-A10 strongly binds gp41 and gp120, most likely preventing HIV-1 attachment/entry through multiple mechanisms. Interestingly, the antiviral activity of SB105-A10 was also detectable in an organ-like structure of human cervicovaginal tissue, in which SB105-A10 inhibited the HIV-1ada R5 strain infection without altering the tissue viability. These results demonstrated the strong antiviral activity of SB105-A10 and suggest a potential microbicide use of this dendrimer to prevent the heterosexual transmission of HIV-1.
Conflict of interest statement
Figures






Similar articles
-
Inhibition of herpes simplex virus type 1 and type 2 infections by peptide-derivatized dendrimers.Antimicrob Agents Chemother. 2011 Jul;55(7):3231-9. doi: 10.1128/AAC.00149-11. Epub 2011 May 16. Antimicrob Agents Chemother. 2011. PMID: 21576438 Free PMC article.
-
Inhibition of human respiratory syncytial virus infectivity by a dendrimeric heparan sulfate-binding peptide.Antimicrob Agents Chemother. 2012 Oct;56(10):5278-88. doi: 10.1128/AAC.00771-12. Epub 2012 Jul 30. Antimicrob Agents Chemother. 2012. PMID: 22850525 Free PMC article.
-
Peptide-derivatized dendrimers inhibit human cytomegalovirus infection by blocking virus binding to cell surface heparan sulfate.Antiviral Res. 2010 Mar;85(3):532-40. doi: 10.1016/j.antiviral.2010.01.003. Epub 2010 Jan 18. Antiviral Res. 2010. PMID: 20083141
-
Mechanistic Studies of Viral Entry: An Overview of Dendrimer-Based Microbicides As Entry Inhibitors Against Both HIV and HSV-2 Overlapped Infections.Med Res Rev. 2017 Jan;37(1):149-179. doi: 10.1002/med.21405. Epub 2016 Aug 12. Med Res Rev. 2017. PMID: 27518199 Review.
-
Potential drug targets on the HIV-1 envelope glycoproteins, gp120 and gp41.Curr Pharm Des. 2003;9(18):1453-62. doi: 10.2174/1381612033454720. Curr Pharm Des. 2003. PMID: 12769725 Review.
Cited by
-
New hope for eradication of HIV from the body: the role of polymeric nanomedicines in HIV/AIDS pharmacotherapy.J Nanobiotechnology. 2014 Mar 22;12:9. doi: 10.1186/1477-3155-12-9. J Nanobiotechnology. 2014. PMID: 24655921 Free PMC article. Review.
-
Advances in Targeting HPV Infection as Potential Alternative Prophylactic Means.Int J Mol Sci. 2021 Feb 23;22(4):2201. doi: 10.3390/ijms22042201. Int J Mol Sci. 2021. PMID: 33672181 Free PMC article. Review.
-
Inhibition of Human Metapneumovirus Binding to Heparan Sulfate Blocks Infection in Human Lung Cells and Airway Tissues.J Virol. 2016 Sep 29;90(20):9237-50. doi: 10.1128/JVI.01362-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27489270 Free PMC article.
-
M48U1 and Tenofovir combination synergistically inhibits HIV infection in activated PBMCs and human cervicovaginal histocultures.Sci Rep. 2017 Feb 1;7:41018. doi: 10.1038/srep41018. Sci Rep. 2017. PMID: 28145455 Free PMC article.
-
Baseline and time-updated factors in preclinical development of anionic dendrimers as successful anti-HIV-1 vaginal microbicides.Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2022 May;14(3):e1774. doi: 10.1002/wnan.1774. Epub 2022 Jan 12. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2022. PMID: 35018739 Free PMC article. Review.
References
-
- WHO (2011) Global HIV/AIDS Response: Epidemic update and health sector progress towards Universal Access Progress report 2011. Available: http://www.who.int/hiv/data/en/Accessed 2013 May10.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

